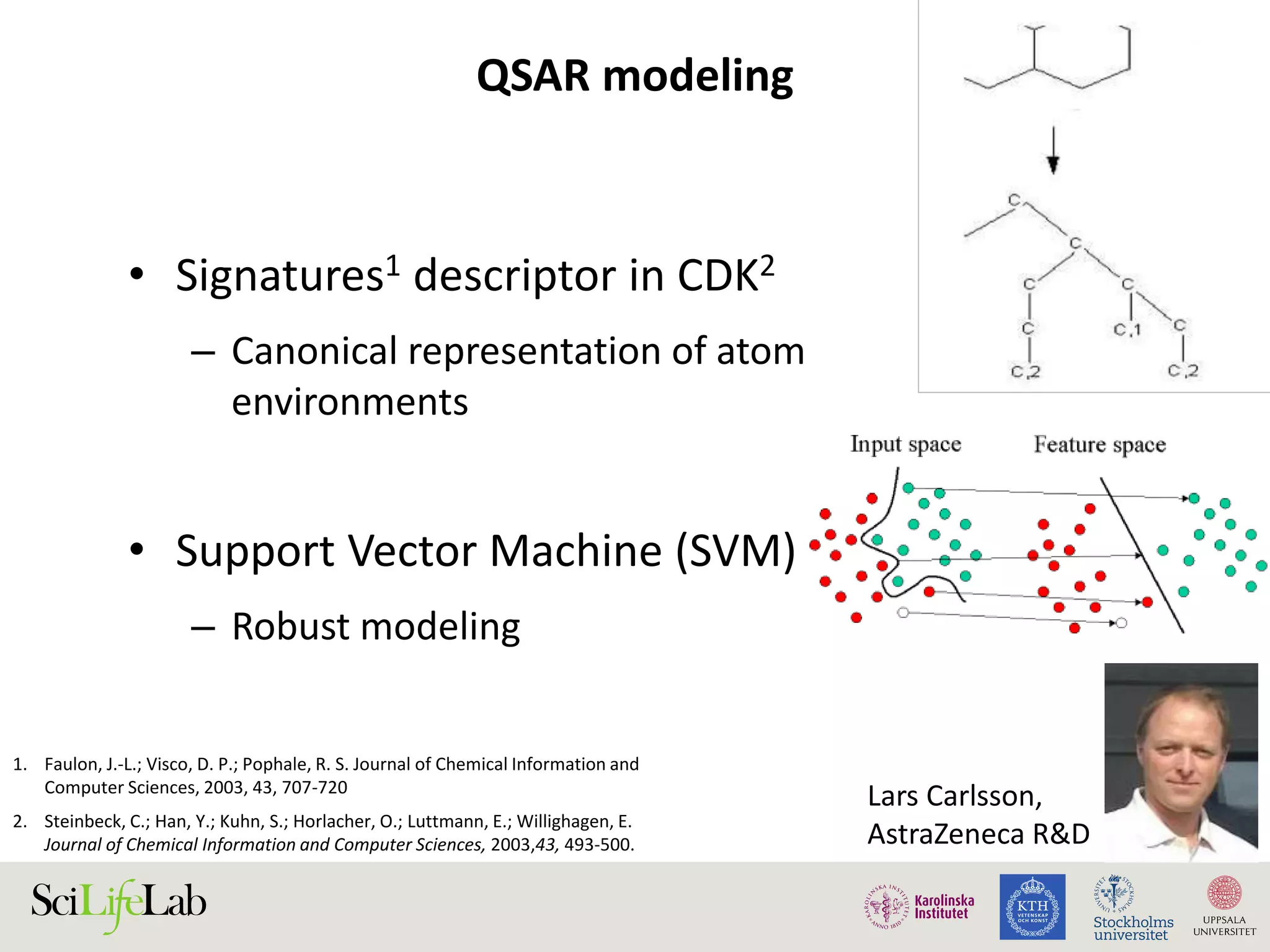
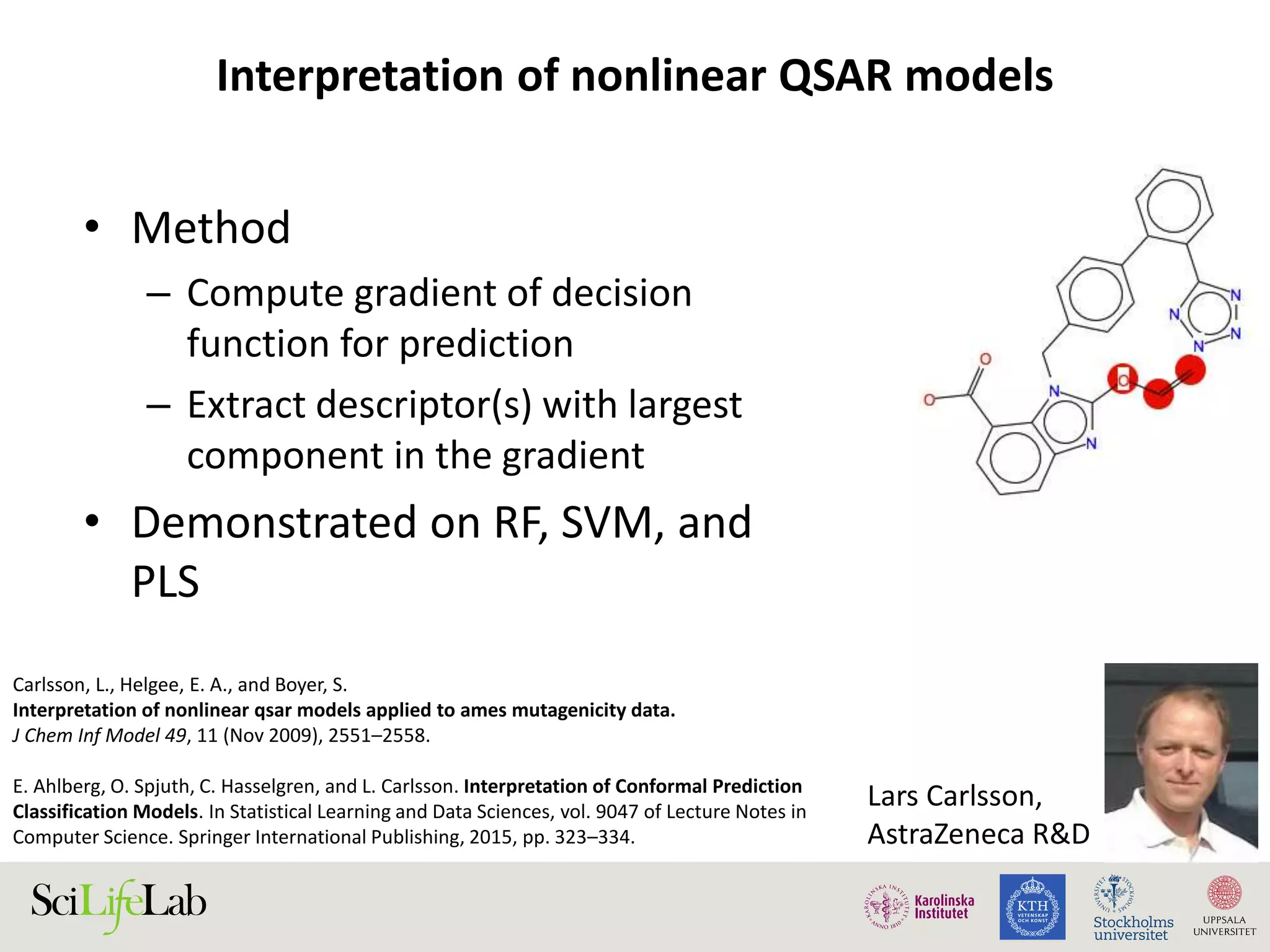
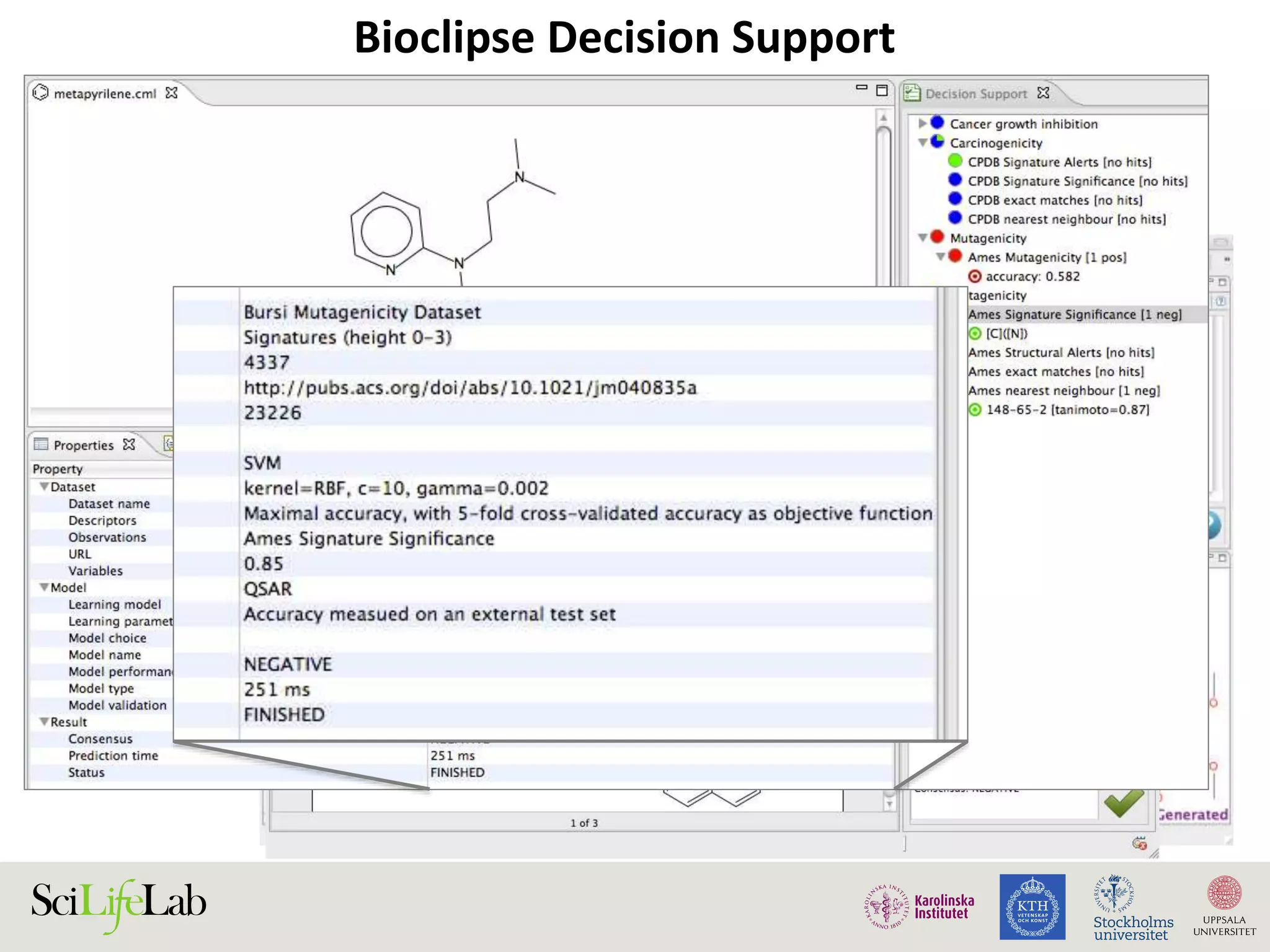
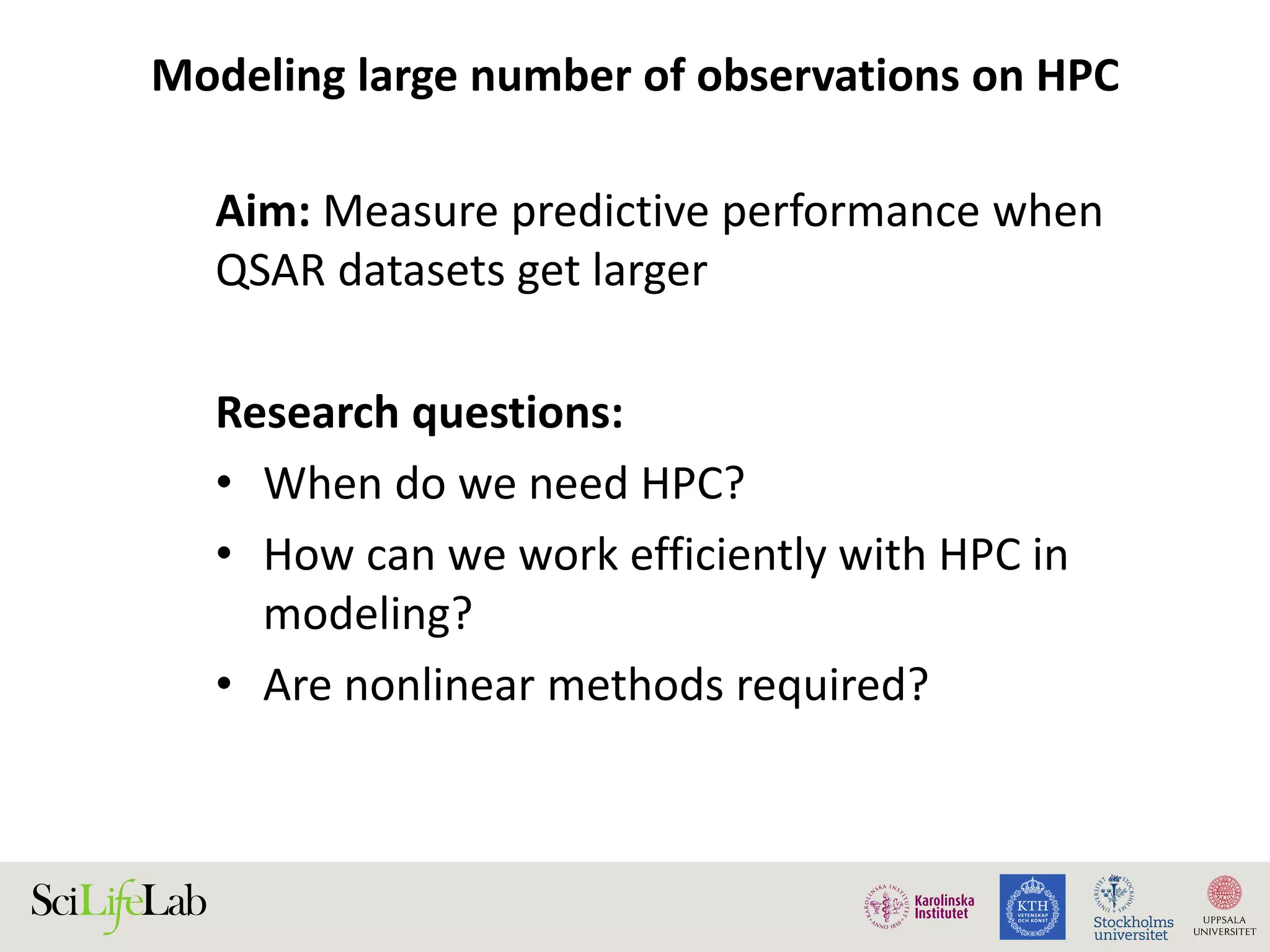
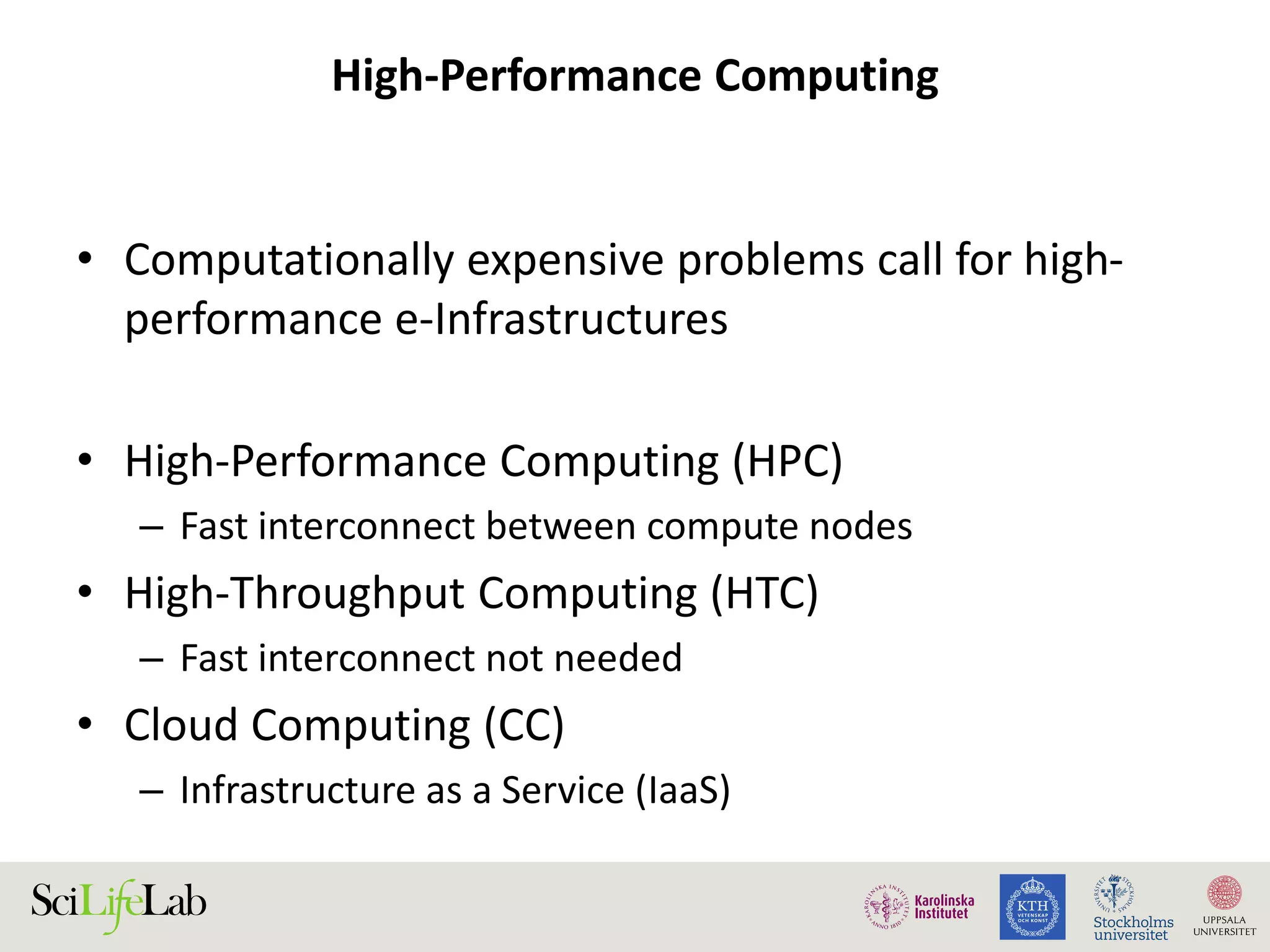
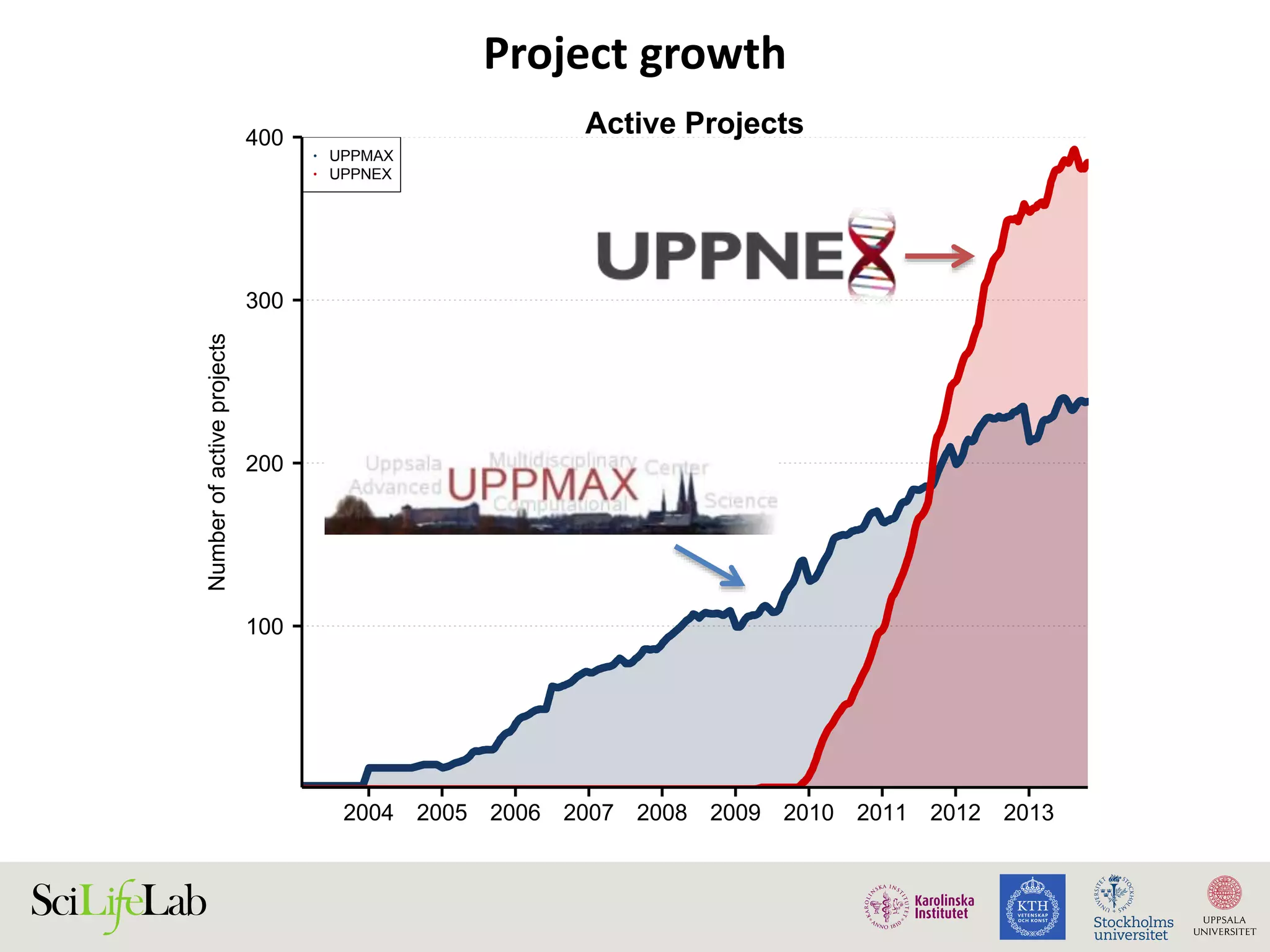
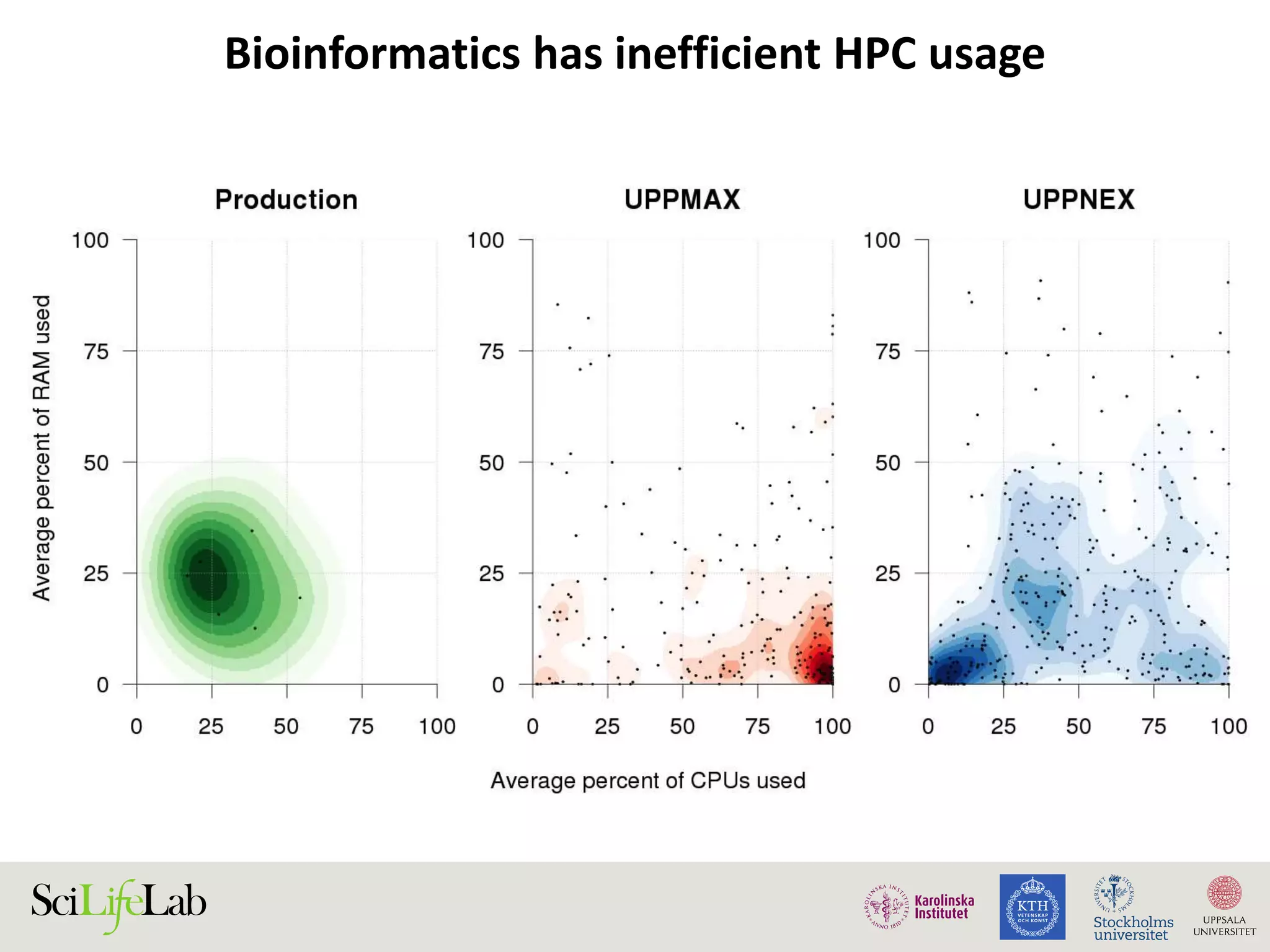
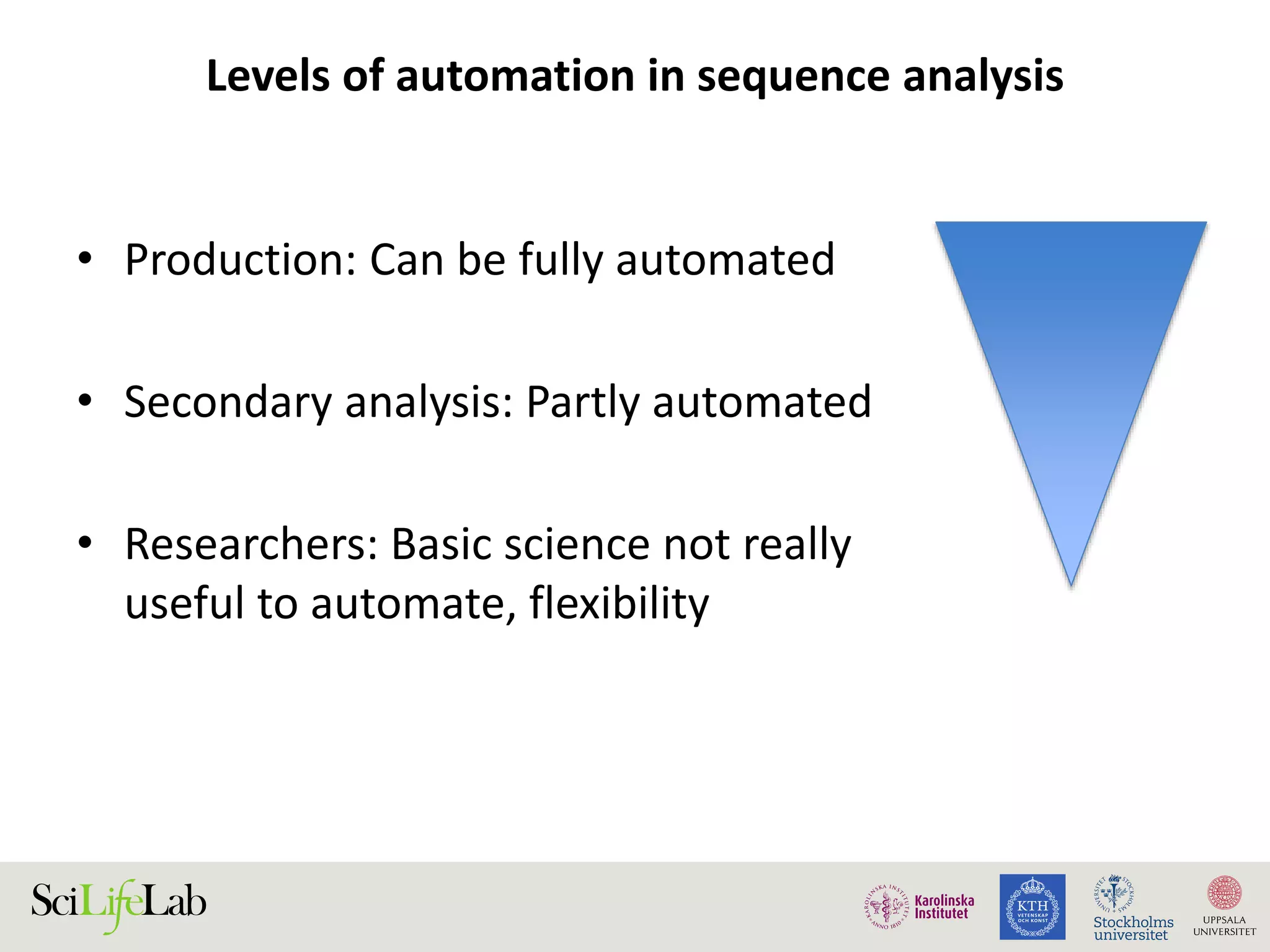
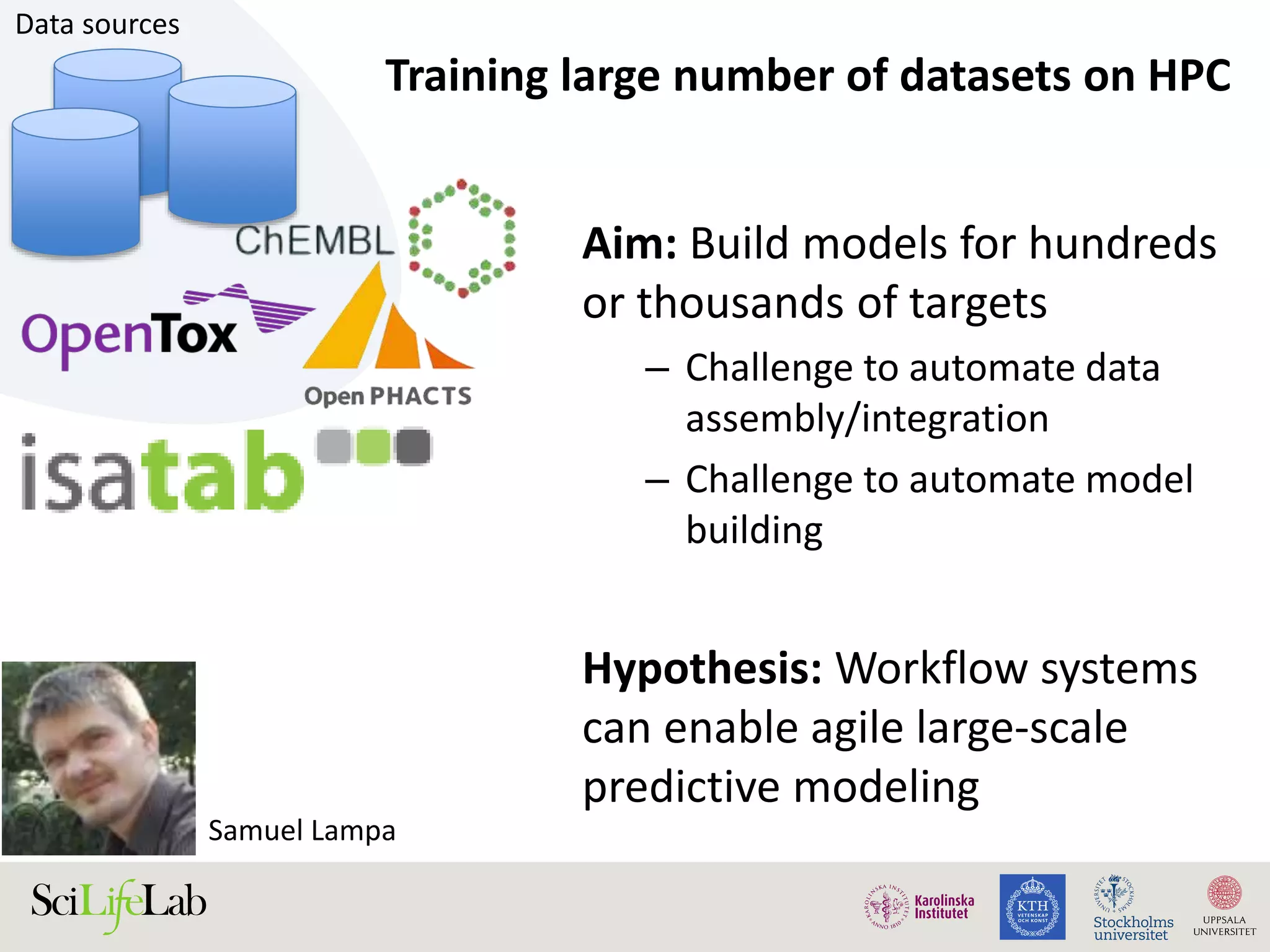
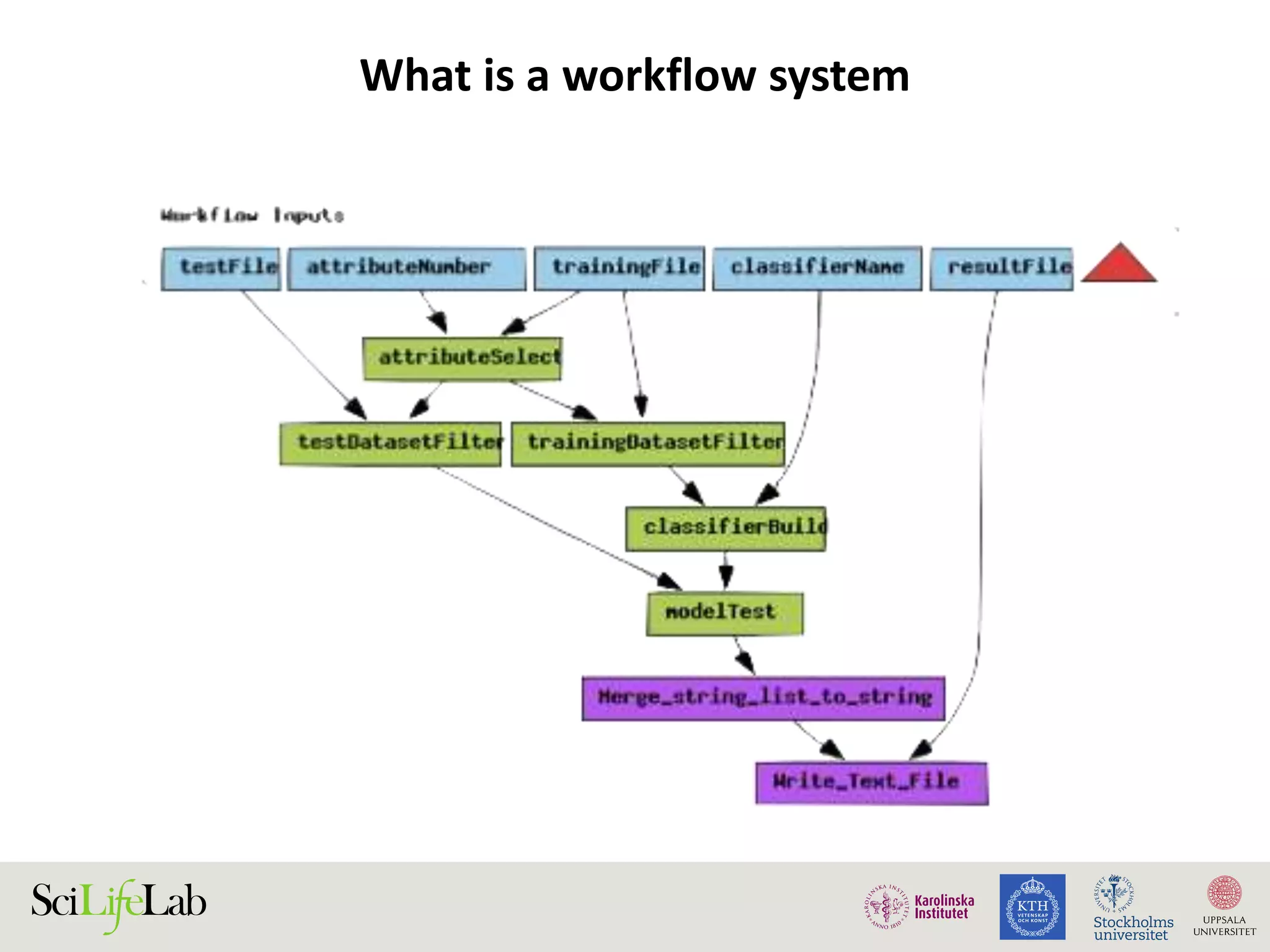
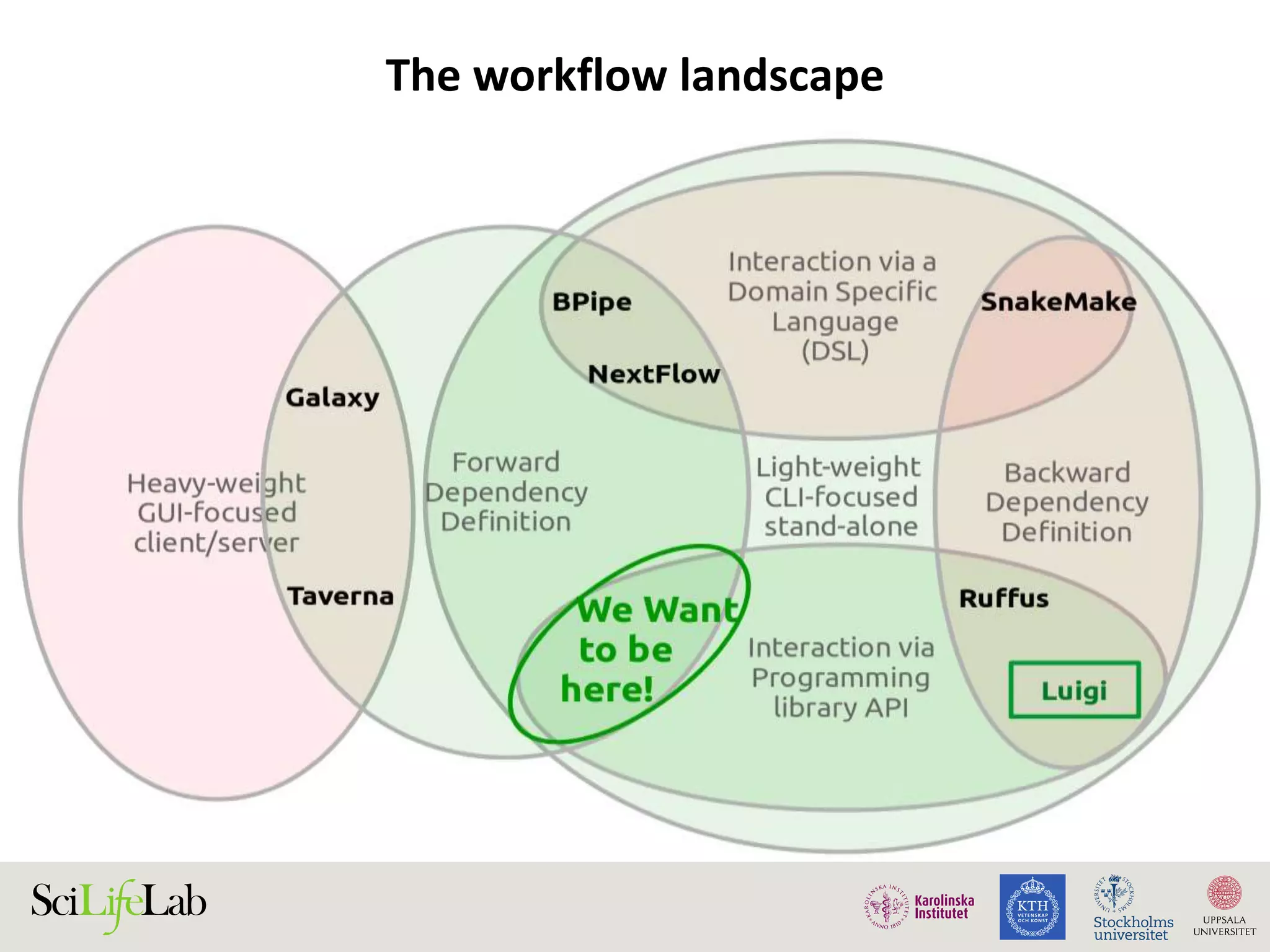
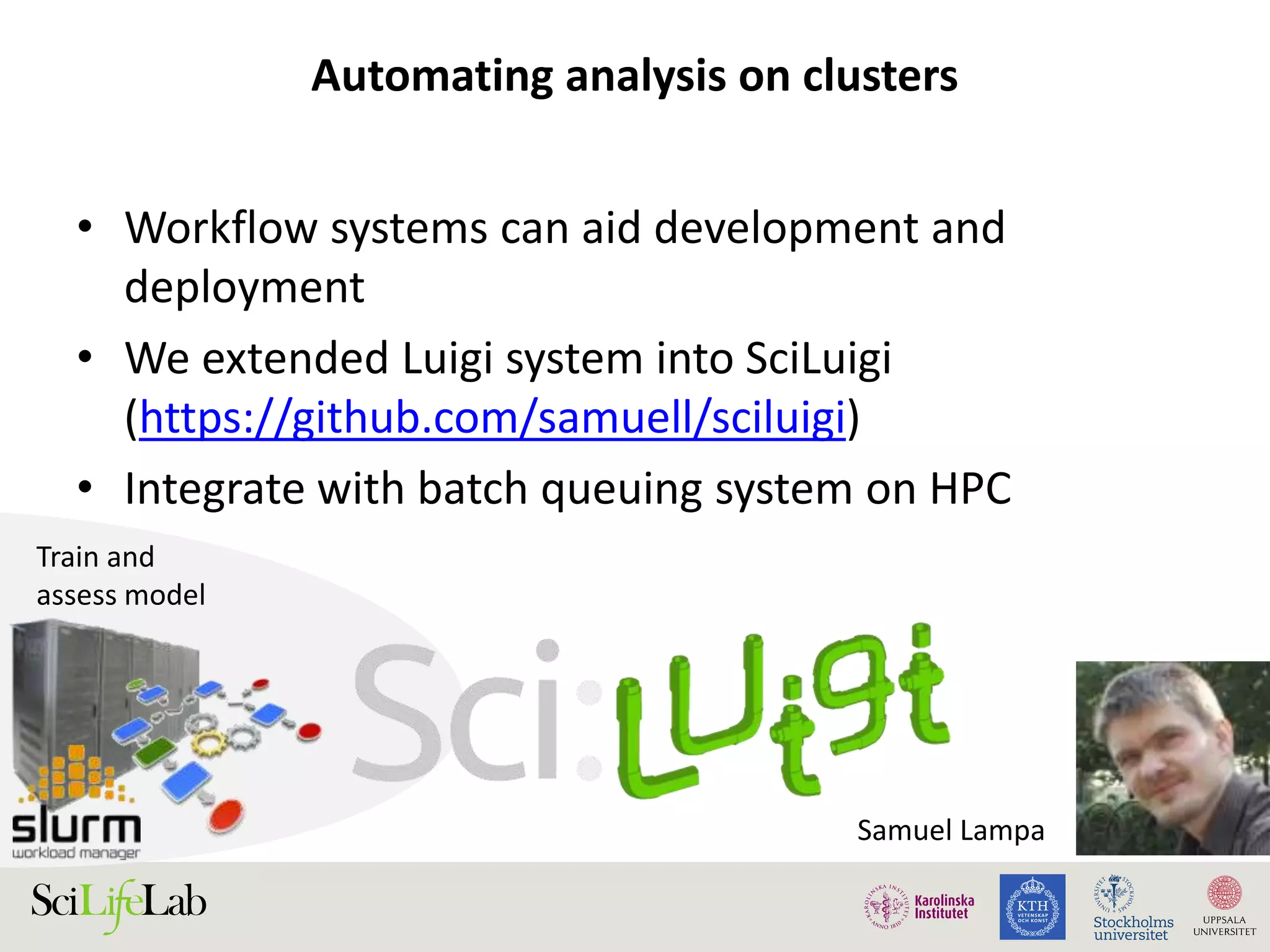
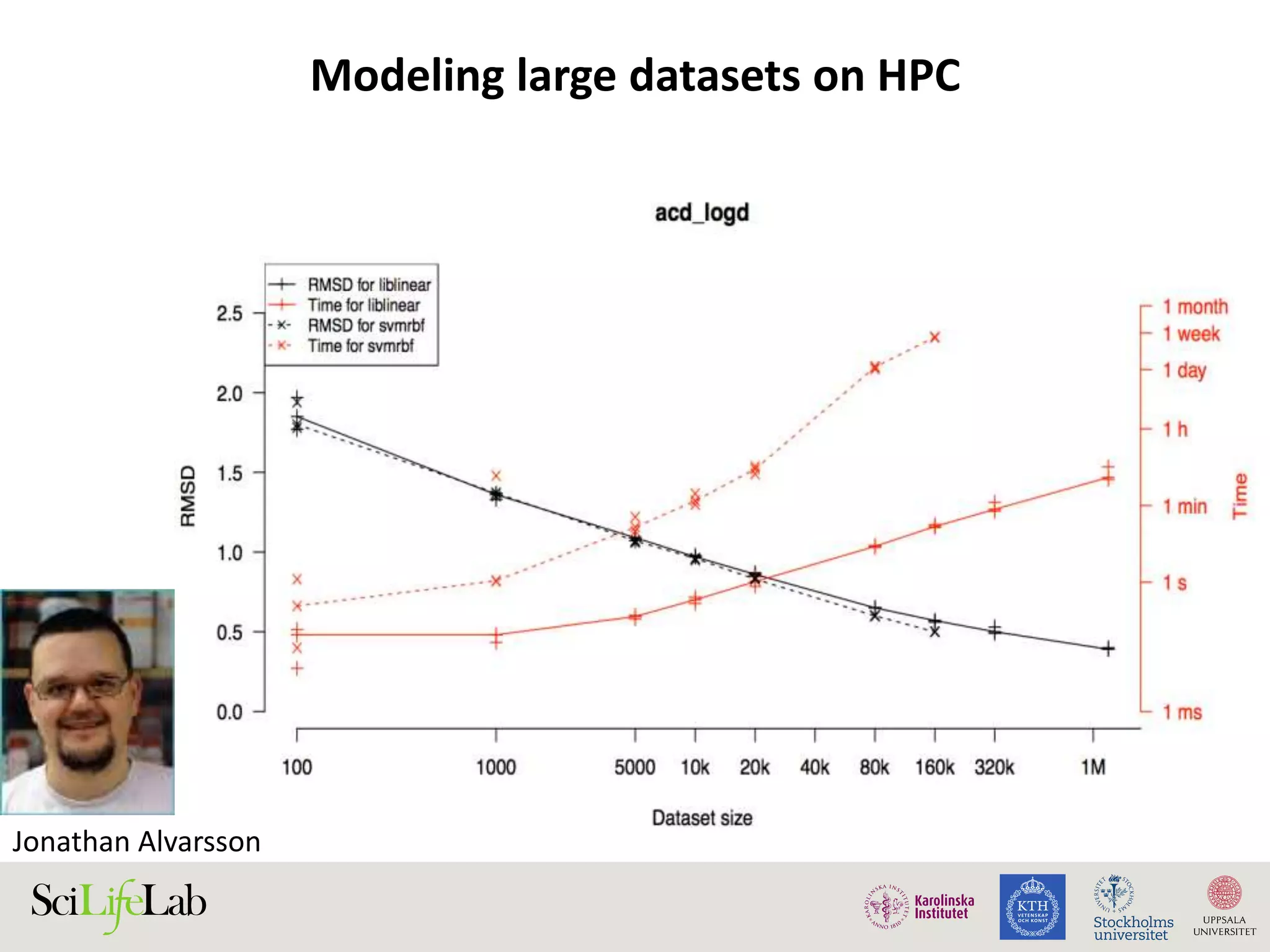
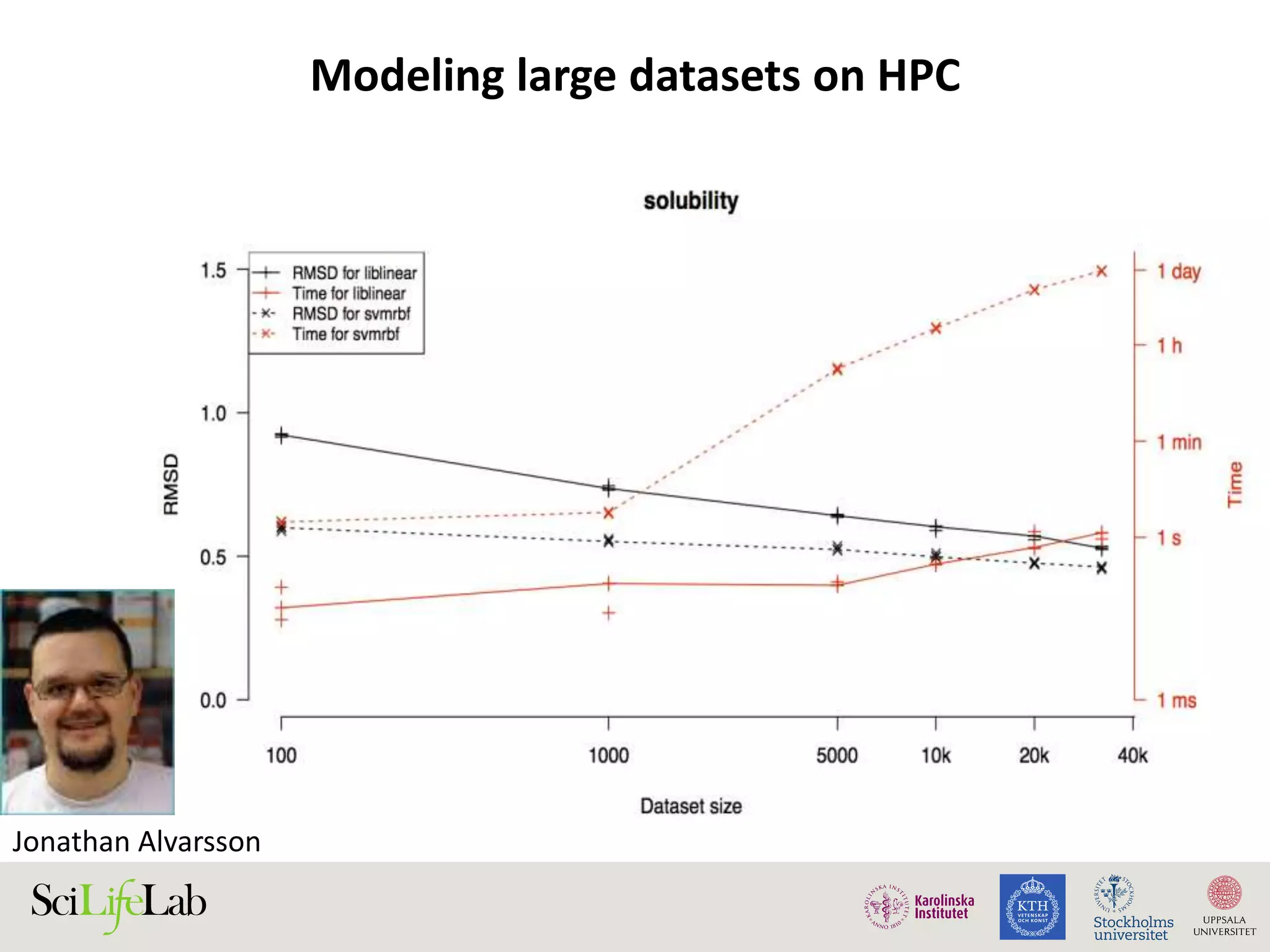
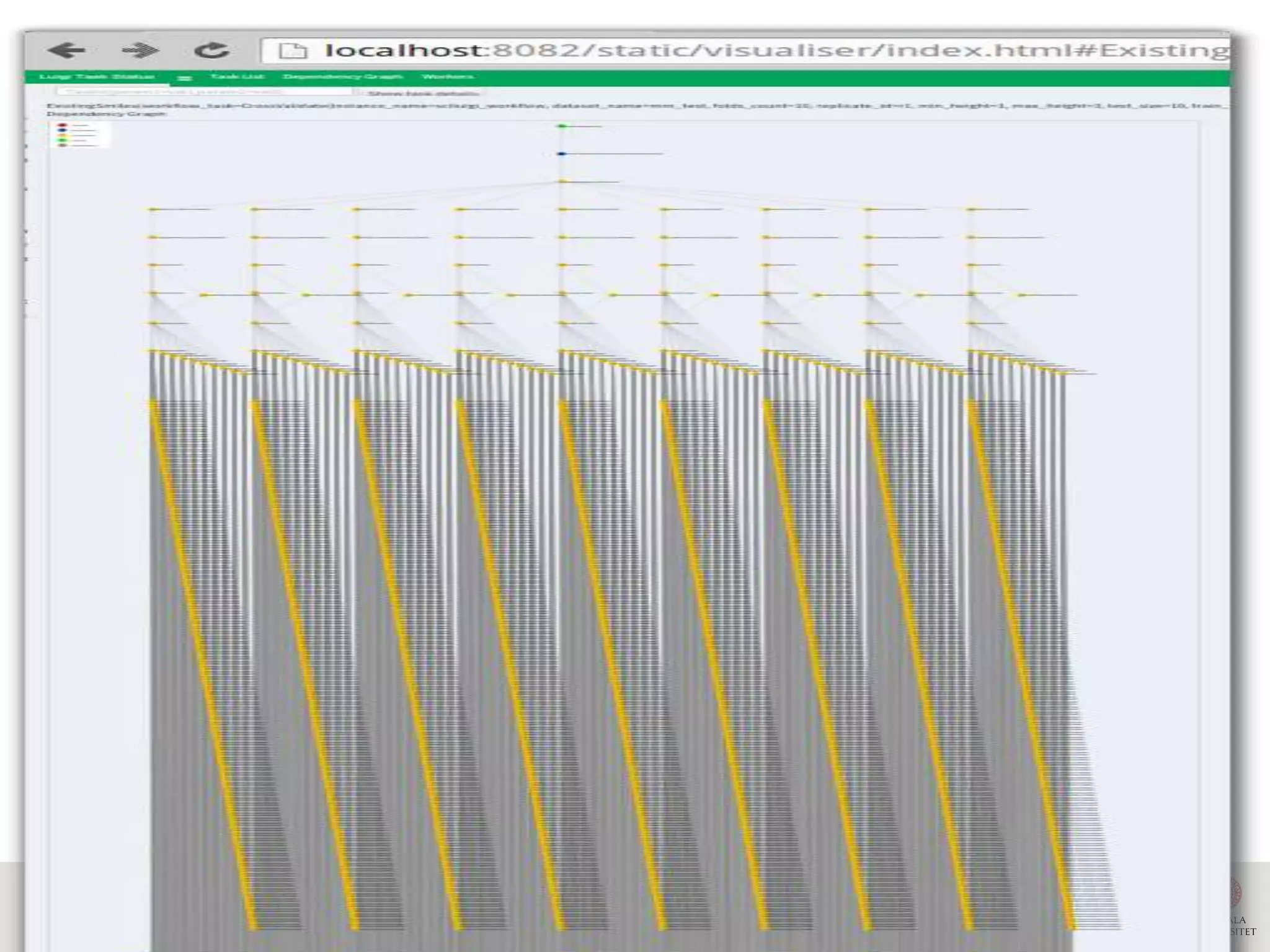
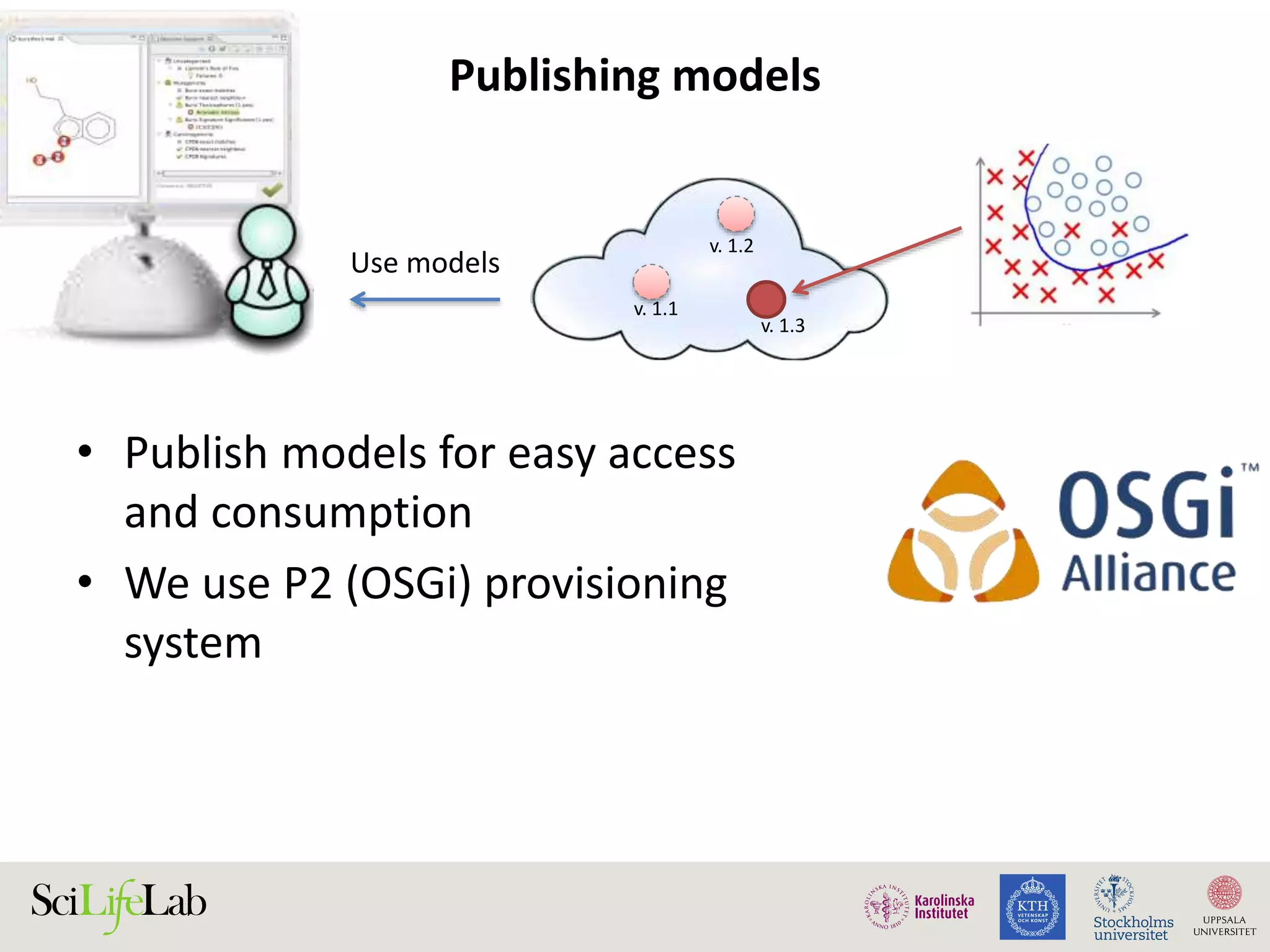
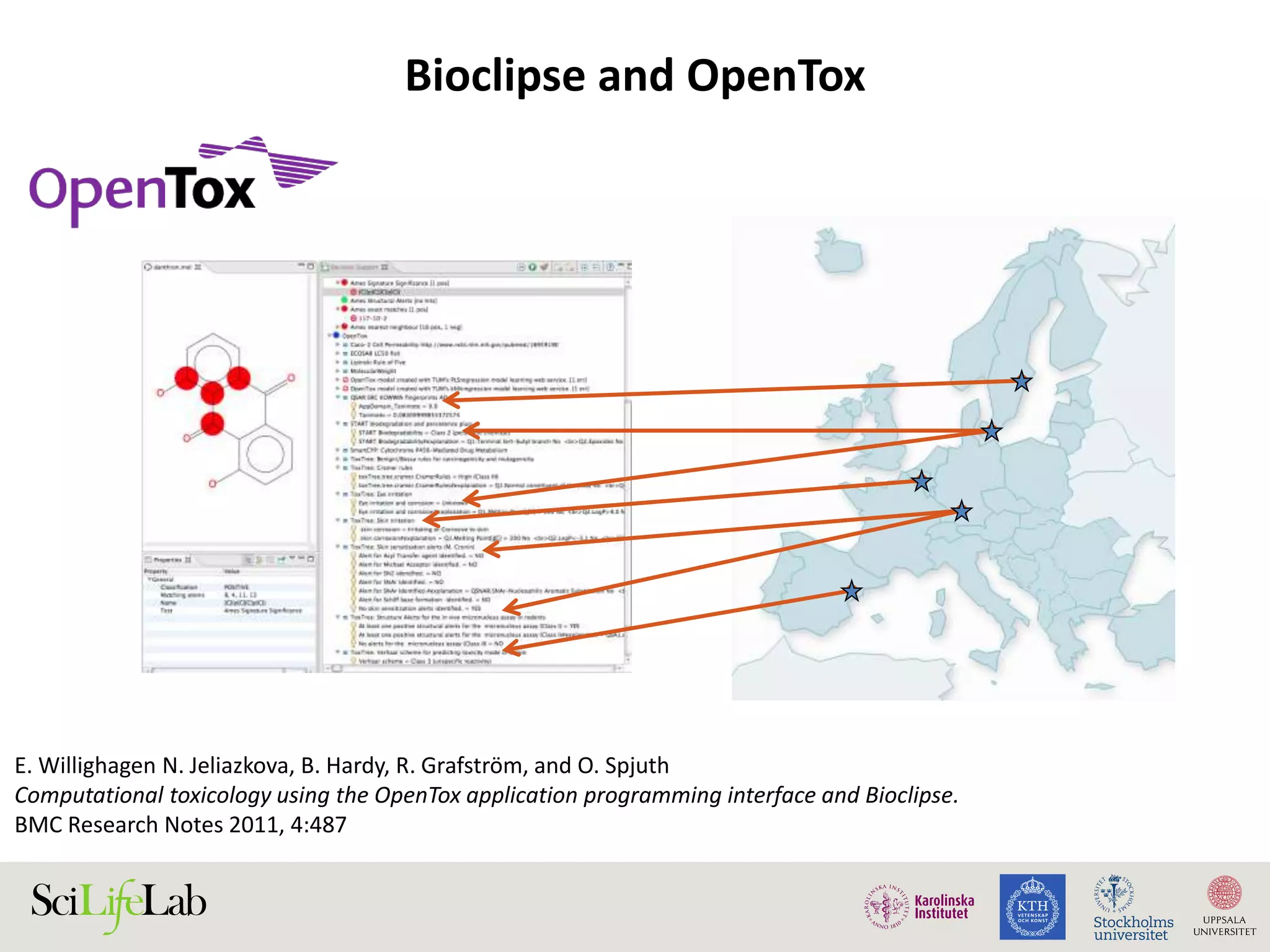
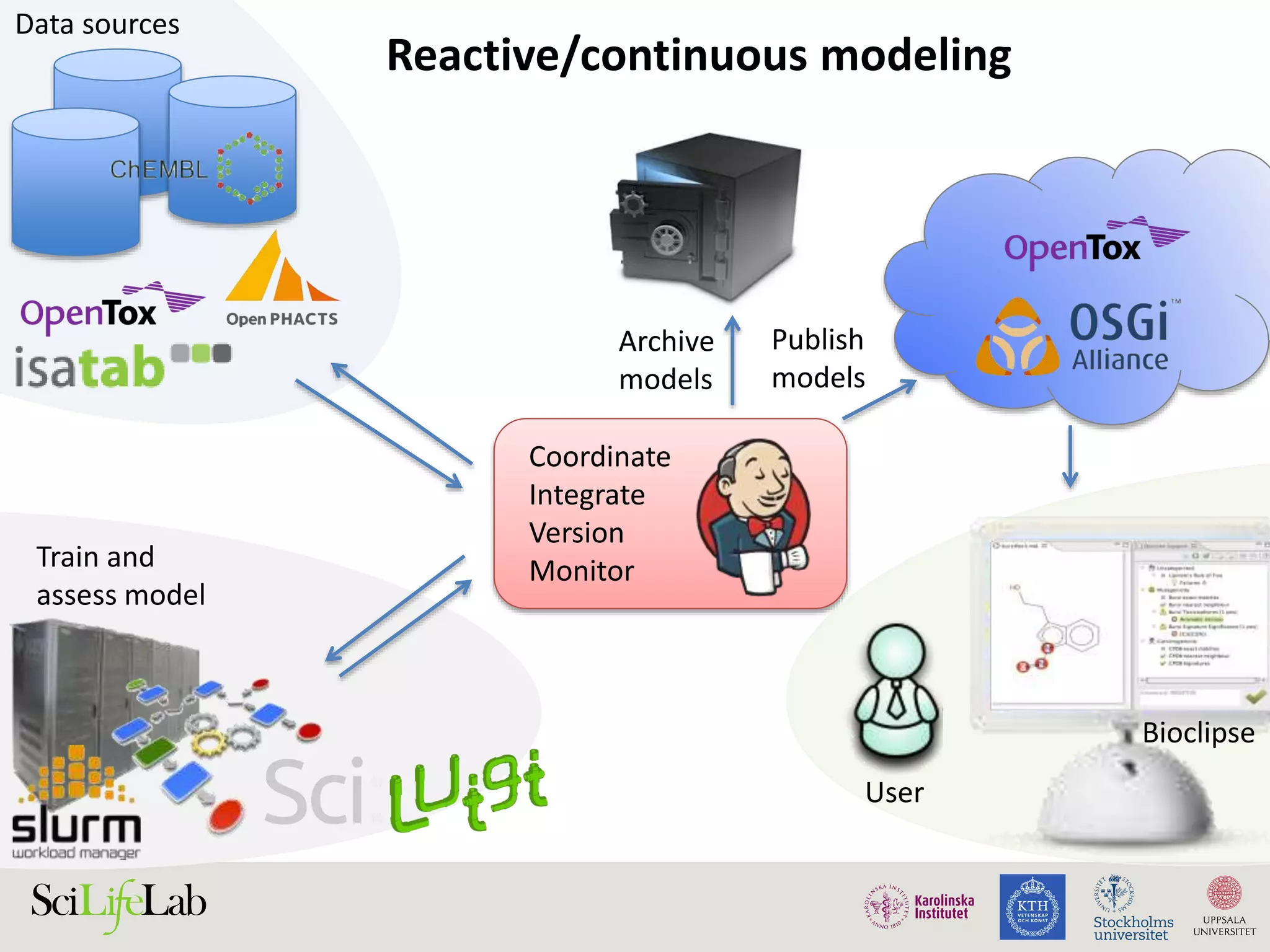
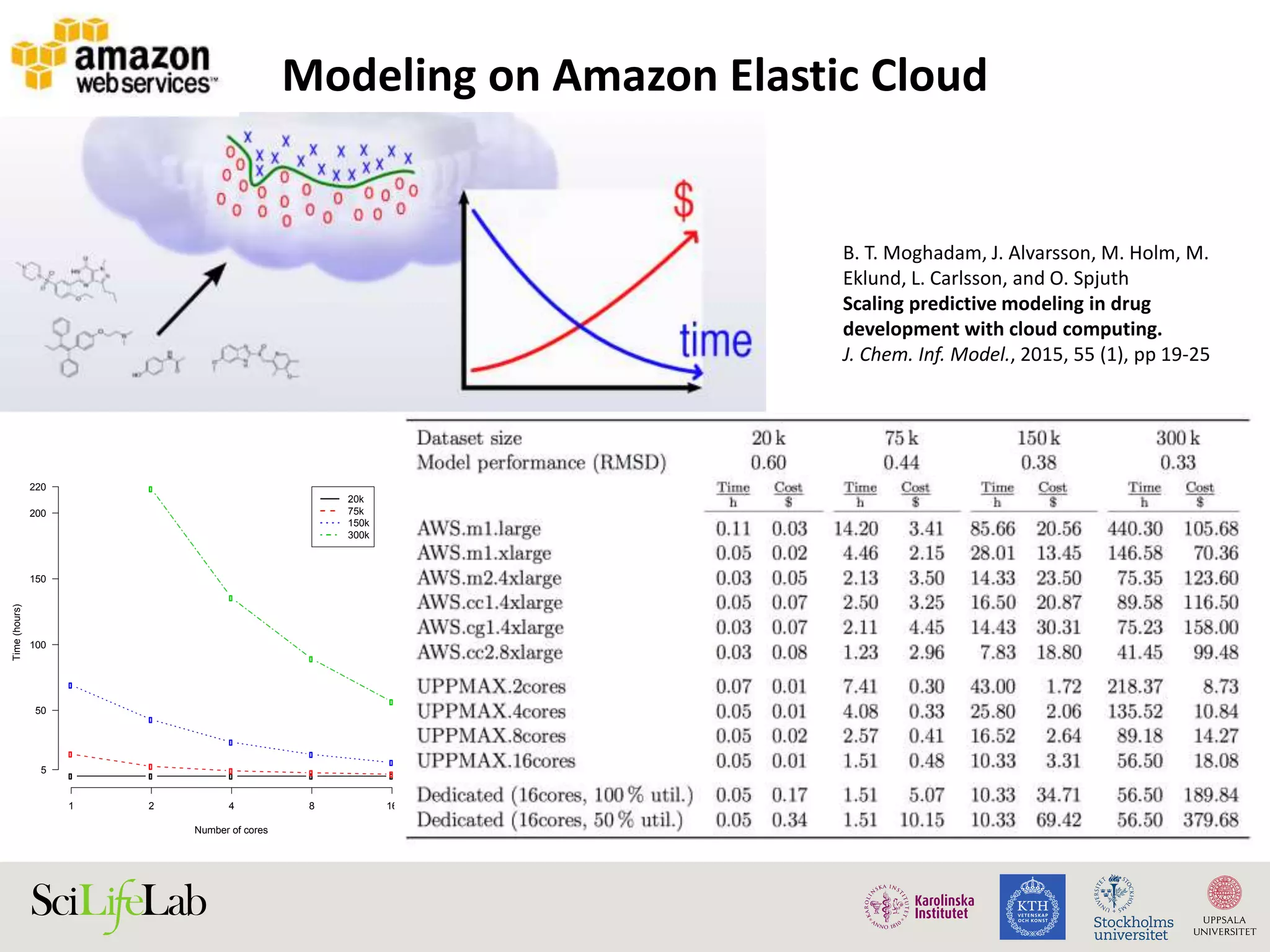
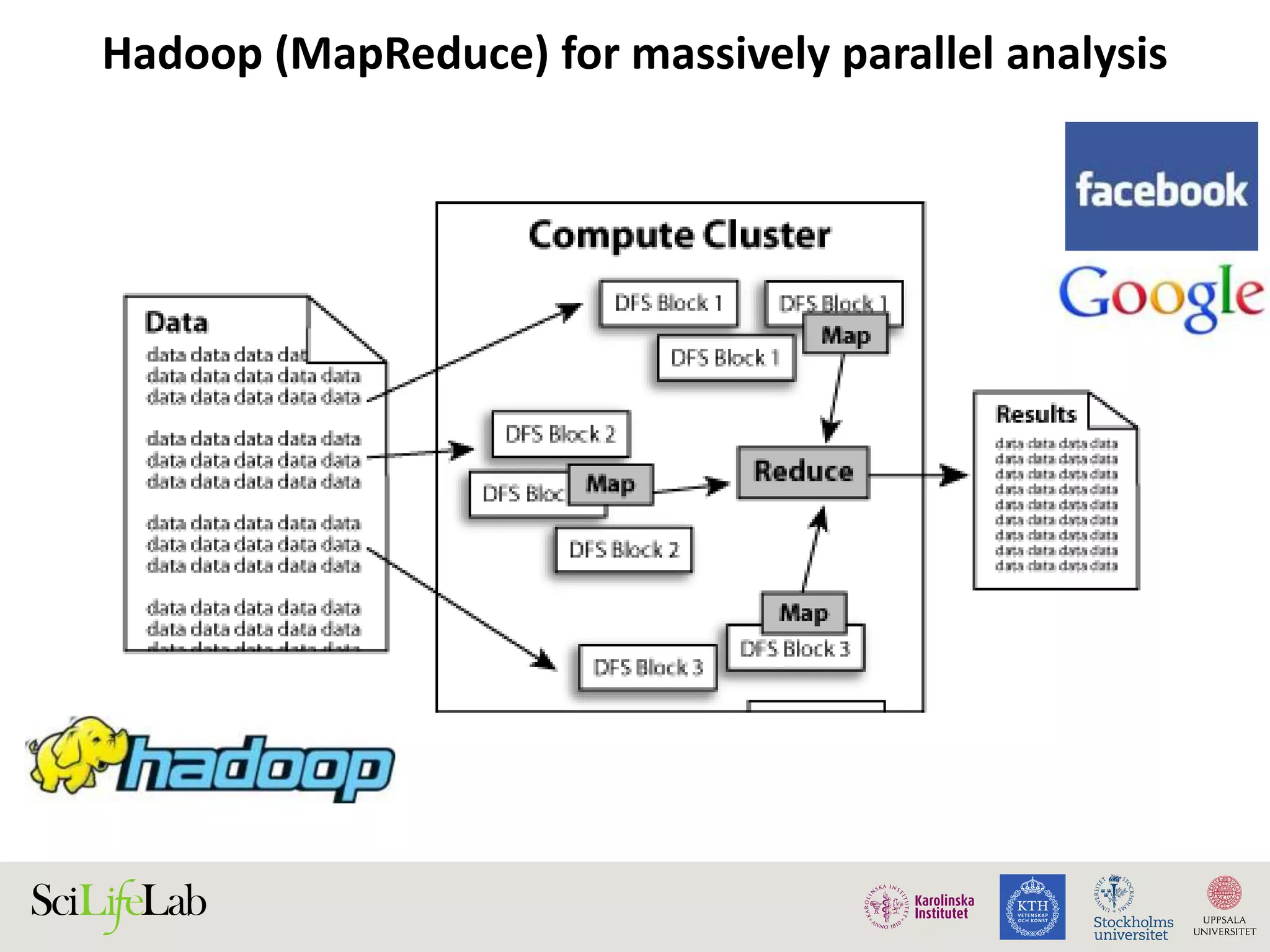
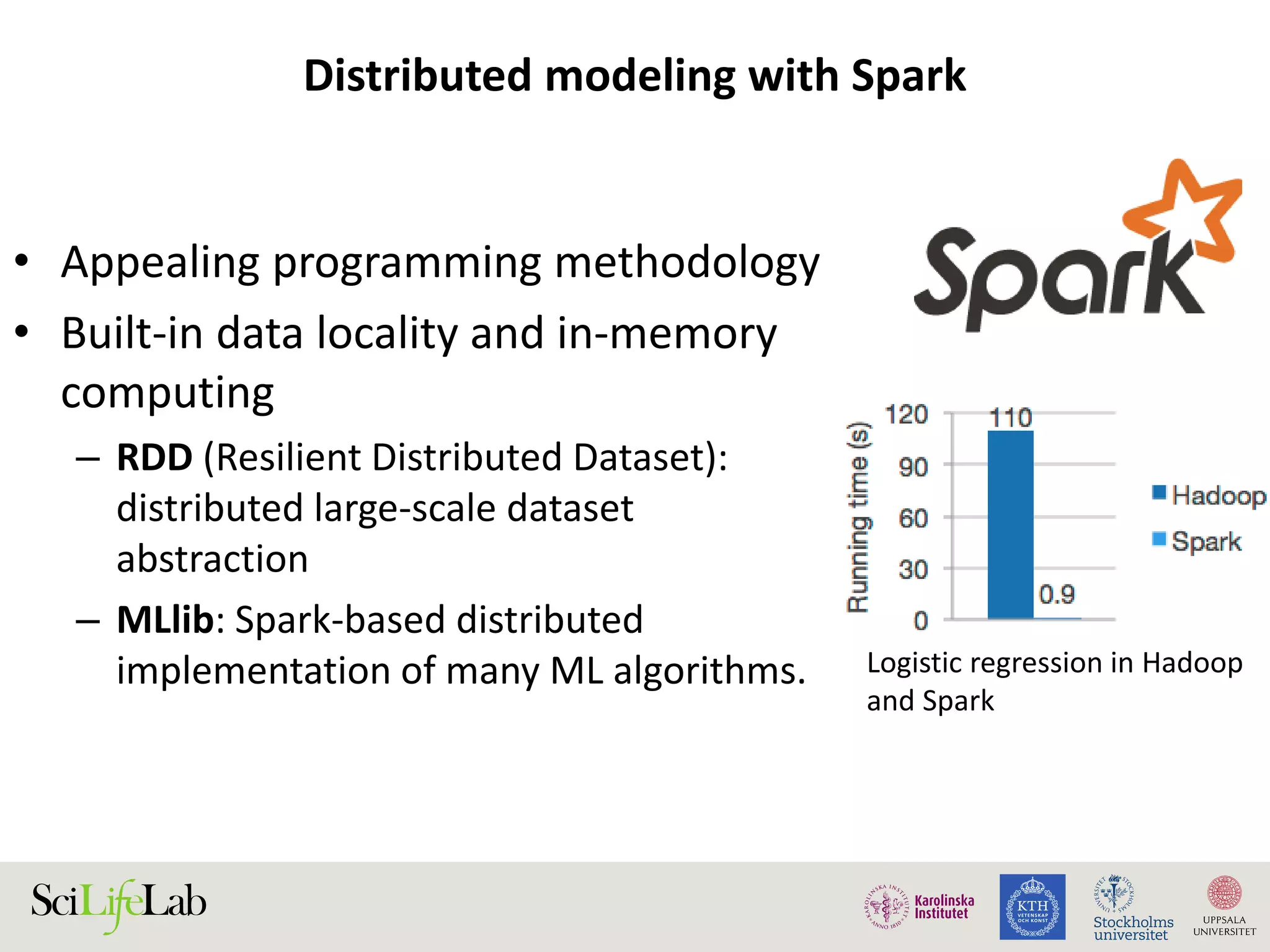
This document discusses continuous modeling and automating model building on high-performance infrastructures. It notes the new challenges of data management, analysis, scaling, and automation posed by high-throughput technologies. The author's research focuses on enabling high-throughput biology through large-scale predictive modeling, evaluating performance, and automating model re-building. Predictive toxicology and pharmacology are becoming data-intensive due to more data sources. The document explores modeling large datasets on high-performance computing infrastructures and whether workflow systems or cloud/Big Data frameworks could improve modeling.