Prokaryotic replication
- 1. Compiled by V. Magendira Mani Assistant Professor, PG & Research Department of Biochemistry, Islamiah College (Autonomous), Vaniyambadi, Vellore District – 6357512, Tamilnadu, India. magendiramani@rediffmail.com ; vinayagam magendiramani@academia.edu
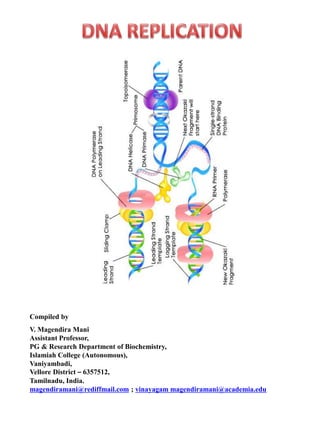
- 2. REPLICATION IN PROKARYOTES Replication is an enzymatic process in which synthesis of a daughter or progeny duplex DNA molecule, identical to the parental duplex DNA occurs. Rate of replication in E.Coli (prokaryotic cell) is 1500 nucleotides per second. To complete replication of whole E.Coli genome it takes 40 minutes. The synthesis or replication of DNA molecule can be divided into three stages I) Initiation (Formation of Replisome) II) Elongation (Initiation of synthesis and elongation) III) Termination
- 3. I) INITIATION The replication begins at a specific initiation point called Ori C point or replicon. (Replicon: It is a unit of the genome in which DNA is replicated; it contains an origin for initiation of replication) It is the point of DNA open up and form open complex leading to the formation of prepriming complex to initiate replication process. The Ori C site consists of 245 basepairs, of which three of 13 basepair sequence are highly conserved in many bacteria and forms the consensus sequences (GATCTNTTNTTTT). Close to OriC site, there are four of 9 basepair sequences each (TTATCCACA).
- 4. The sequence of reactions in the initiation process is as follows: a) Dna A protein recognizes and binds up to four 9 bp repeats in Ori C to form a complex of negatively supercoiled Ori C DNA wrapped around a central core of Dna A protein monomers. This process requires the presence of the histone like HU or 1 HC proteins to facility DNA bending. b) Once the four 9 bp repeats are occupied 20 – 40 additional Dna A monomers bind, so that entire Ori C region is complexes with Dna A protein. c) The resulting complex resembles a nucleosome with negatively supercoiled Ori C DNA wrapped around a DNA core.
- 6. d) HU, a histone like protein prevents nonspecific initiation at sites other than the Ori C. e) Dna A protein subunits then successively melt three tandemly repeated 13 bp segments in the presence of ATP, which results in the formation of 45 bp open complex. f) The Dna A protein then guides a Dna B - Dna C complex into the melted region to form a so called prepriming complex. The Dna C is subsequently released. Dna B further unwinds open complex to form prepriming complex. g) DNA gyrase, single stranded binding protein (SSB), Rep protein and Helicase - II are bound to prepriming complex and now complex is called as priming complex.
- 8. h) In the presence of gyrase and SSB, helicases further unwinds the DNA in both directions so as to permit entry of primase and RNA polymerase. Then RNA polymerase forms primer for leading strand synthesis while primase in the form of primosome synthesis primer for lagging strand synthesis. f) To the above complex, DNA polymerase - III will bind and forms replisome. REPLISOME: It is the multiprotein structure that assembles at the bacterial replicating fork to undertake synthesis of DNA. It contains DNA polymerase and other enzymes.
- 9. II) ELONGATION Now the stage is set for the initiation of synthesis and the elongation to proceed. But this occurs in two mechanistically different pathways in the 5'-->3' template strand and 3'-->5' template strand. Initiation of synthesis and Elongation on the 5'-->3' template (If replication fork moves in 3'-->5' direction). Synthesis of leading strand The DNA daughter strand that is synthesized continuously on 5'-->3' template is called leading strand. Leading strand synthesis begins with the synthesis of RNA primer (10 -60 nucleotides) by primase (DNA g protein)
- 10. DNA pol-III synthesizes DNA by adding 5'-P of deoxynucleotide to 3'-OH group of the already presenting fragment. Thus chain grows in 5'-->3' direction. The reaction catalyzed by DNA pol-III is very fast. The enzyme is much more active than DNA pol - I and can add 9000 nucleotides per minute at 37*C. The RNA primer that was initially added by RNA polymerase is degraded by RNase.
- 11. Synthesis of lagging strand Initiation of synthesis and Elongation on 3'-->5' template when fork moves in 3'-->5' direction The daughter DNA strand which is synthesized in discontinuous complex fashion on the 3'-->5' template is called lagging strand. It occurs in the following steps: i) Synthesis of Okazaki fragment: To the RNA primer synthesized by primosome, 1000-2000 nucleotides are added by DNA pol-III to synthesis Okazaki fragments. ii) Excision of RNA primer: When the Okazaki fragment synthesis was completed up to RNA primer, then RNA primer was removed by DNA pol - I using its 5'-->3' exonuclease activity.
- 12. iii) Filling the gap (Nick translation) The gap created by the removal of primer, is filled up by DNA pol - I using the 3'-OH of nearby Okazaki fragment by its polymerizing activity. iv) Joining of Okazaki fragment: (Nick sealing) Finally, the nick existing between the fragments are sealed by DNA ligase which catalyze the formation of phosphodiester bond between a 3'-OH at the end of one strand and a 5' - phosphate at the other end of another fragment. The enzyme requires NAD for during this reaction.
- 14. III) TERMINATION: Termination occurs when the two replicating forks meet each other on the opposite side of circular E.Coli DNA. Termination sites like A, B, C, D, E and F are found to present in DNA. Of these sites, Ter A terminates the counter clockwise moving fork while ter C terminates the clockwise moving forks. The other sites are backup sites. Termination at these sites are possible because, at these sites tus protein (Termination utilizing substance) will bound to Dna B protein and inhibits its helicase activity. And Dna B protein released and termination result. After the complete synthesis, two duplex DNA are found to be catenated (knotted). This catenation removed by the action of topoisomerase. Finally, from single parental duplex DNA, two progeny duplex DNA synthesized.
- 15. REGULATION OF PROKARYOTIC REPLICATION Especially initiation of replication is regulated. Dna A protein when available in high concentration then ratio of DNA to cell mass is quiet high but at low Dna A concentration, the ratio found to be low. This shows that Dna A protein regulates the initiation of replication. The sequence most commonly methylated in E.Coli is GATC including in three of 13mer sequence. Thus, the observation that E.Coli defective in the GATC methylation enzyme are very inefficiently replicated, suggests that the DNA replication trigger also responds to the level of Ori C methylation.
- 16. ALL THE BEST By VMM V. Magendira Mani Assistant Professor, PG & Research Department of Biochemistry, Islamiah College (Autonomous), Vaniyambadi, Vellore District – 6357512, Tamilnadu, India. magendiramani@rediffmail.com ; vinayagam magendiramani@academia.edu https://tvuni.academia.edu/mvinayagam