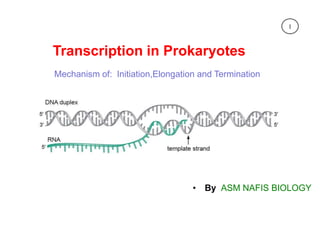
Transcription in prokaryotes.
- 1. Transcription in Prokaryotes Mechanism of: Initiation,Elongation and Termination • By ASM NAFIS BIOLOGY 1
- 2. Objectives • Understand the basic mechanism of Transcription. • Know the process of polymerase in Transcription • Know the function of promoter elements and its associating proteins. • Know the proof reading mechanism • Know the Transcription cycle(initiation,elongation ,termination). • Understand the process of termination (by independently or by involvement of rho protein). 2 ASM NAFIS BIOLOGY
- 4. Bacterial promoters There are several flavors of promoters and recruit RNAP to promoter DNA 4 ASM NAFIS BIOLOGY
- 5. RNA polymerases 5 ASM NAFIS BIOLOGY
- 7. RNA Polymerase and its Promoter intraction 7 ASM NAFIS BIOLOGY
- 8. Challenges in initiating transcription 1.Job of RNAP is to make transcripts—has to elongate approximately without regard to sequence; and the enzyme is specialized in this way; starting trx at defined positions requires specific recognition; ortohogonal to elongation—whole new apparatus to do this. 2.The initiating enzyme must not only recognize DNA but open strands template strand is available to guide transcription. Contours of that process just being unraveled now—Discussion paper 3.How does the enzyme leave the promoter, when there are all these DNA – protein interactions to bind it to the promoter? Abortive initiation and its relationship to initiation factors is key 8 ASM NAFIS BIOLOGY
- 9. The Initiating Form of RNA Polymerase 9 ASM NAFIS BIOLOGY
- 10. Is the -10 promoter element recognized as Duplex or SS DNA? -10 logo-35 logo Helix-turn-helix in Domain 4 Recognizes -35 as duplex DNA Recognition of the prokaryotic promoter Domain 2 recognizes - 10 as duplex DNA 11 ASM NAFIS BIOLOGY
- 11. ‘holoenzyme’ '2 7 + 7 ‘core’ } Can begin transcription on promoters and can elongate } Can elongate but cannot begin transcription at promoters factor is required for bacterial RNA polymerase to initiate transcription on promoters '2 12 ASM NAFIS BIOLOGY
- 12. Initiating RNAP must open DNA to permit transcription: Formation of the open complex 13 ASM NAFIS BIOLOGY
- 13. is positioned for DNA recognition 14 ASM NAFIS BIOLOGY
- 14. Undergoes a large conformational change upon binding to RNA polymerase Free doesn’t bind DNA in holoenzyme positioned for DNA recognitionSorenson; 2006 15ASM NAFIS BIOLOGY
- 15. is positioned to affect key activities of RNA polymerase 16 ASM NAFIS BIOLOGY
- 16. Fig-Recognition and melting of -10 element by sigma region 2 Ref ch12 watson 17 ASM NAFIS BIOLOGY
- 17. Remember that sigma linker 3-4 and the b reader of TFB are in the RNA exit channel, providing a barrier to chain extension and presumably blocking RNA exit ( not enough room for both sigma linker and RNA) It had been established that sigma domain 4 dissociates from RNAP first. That brings us to this very curious process carried out by all RNA –abortive iniitation—many short RNA chains made prior to extension. Tell you about recent work of ebright to understand the mechanistic basis of this process, and the insights it provided into the role of abortive initiation in promoter excape 18 ASM NAFIS BIOLOGY
- 18. Steps in transcription initiation 19 ASM NAFIS BIOLOGY
- 19. Promoter escape and Abortive Initiation during abortive initiation, RNAP synthesizes many short transcripts, but reinitiates rapidly. How can the active site of RNAP move forward along the DNA while maintaining promoter contact? 2 ASM NAFIS BIOLOGY
- 20. Three models for Abortive initiation #1 Predicts expansion and contraction of RNAP Predicts expansion and contraction of DNA Predicts movement of both the RNAP leading and trailing edge relative to DNA #2 #3 23 ASM NAFIS BIOLOGY
- 21. • One model postulates that the RNAP molecule makes transient downstream excursions on the template, briefly breaking its bonds with the promoter, until the short RNA is released, and then the enzyme diffuses back to the promoter Such a model is not easily reconciled with bulk footprinting data, which suggest that the abortive initiation process results from an inability of RNAP to break its promoter contacts. These observations led Straney & Crothers (to propose that the energy required to break free of the promoter might be somehow stored in a “stressed intermediate” and that abortive initiation was a consequence of this energy not being used productively. • One particular instance of this concept, the “inchworming” model, postulates that flexible elements inside RNAP might allow the active center to move forward transiently with respect to the upstream face during synthesis, storing up energy like a stretched spring that retracts upon aborted synthesis. In a third model, the flexible element that stores the energy ultimately used for promoter escape lies not in RNAP but in the single-stranded DNA of the transcription bubble and its interactions with the enzyme. • In this scrunching model, RNAP functions more or less as a rigid body. The downstream DNA is pulled progressively into the enzyme with each nucleotide addition cycle, producing a scrunched form within the enzyme footprint .Abortive RNA transcripts lead to the release of the scrunched DNA, which is then extruded out the downstream face of RNAP (1, 56–58), only to be reeled in again upon further RNA synthesis. 24 ASM NAFIS BIOLOGY
- 28. summery 1. Multisubunit RNA polymerases are conserved among all organism and cannot initiate transcription on their own. In bacteria 70 is required to initiate transcription at most promoters. Among other functions, it recognizes the key features of most bacterial promoters, the -10 and -35 sequences. 2. E. coli RNA polymerase holoenzyme, (core + ) finds promoter sequences by sliding along DNA and by transfer from one DNA segment to another. This behavior greatly speeds up the search for specific DNA sequences in the cell and probably applies to all sequence-specific DNA-binding proteins. 3. Transcription initiation proceeds through a series of structural changes in RNA polymerase, 70 and DNA. 31 ASM NAFIS BIOLOGY
- 29. • A key intermediate in E. coli transcription initiation is the open complex, in which the RNA polymerase holoenzyme is bound at the promoter and ~12 bp of DNA are unwound at the transcription startpoint. Open complex formation does not require nucleoside triphosphates. Its presence can be monitored by a variety of biochemical and structural techniques. • Recognition of the -10 element of the promoter DNA is coupled with strand separation • When the open complex is given NTPs, it begins the ‘abortive initiation’ phase, in which RNA chains of 5-10 nucleotides are continually synthesized and released. • Through a “DNA scrunching” mechanism the energy captured, during synthesis of one of these short transcripts eventually breaks the enzyme loose from its tight connection to the promoter DNA, and it begins the elongation phase. • Aspects of the mechanism of initiation are likely to be conserved . . in eukaryotic RNA polymerase 32 ASM NAFIS BIOLOGY
- 30. Refrences • Young BA, Gruber TM, Gross CA. (2004) Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 303:1382- 1384 • *Kapanidis, AN, Margeat, E, Ho, SO,.Ebright, RH. (2006) Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 314:1144-1147. • Revyakin A, Liu C, Ebright RH, Strick TR (2006) Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 314: 1139-43. • Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA (2002). Structural basis of transcription initiation: an RNA polymerase holoenzyme- DNA complex. Science. 296:1285-90. • Chpter 12 of mol bio 6th edition (2008) by Watson and bekar. ASM NAFIS BIOLOGY
- 31. Discussion Paper **Feklistov A and Darst, SA (2011) Structural basis for Promoter -10 Element recognition by the Bacterial RNA Polymerase Subunit. Cell 147: 1257 – 1269 Accompanying preview: Liu X, Bushnell DA and Kornberg RD ( 2011) Lock and Key to Transcription: –DNA Interaction. Cell: 147: 1218-1219 ***Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. (2004) DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 6:311-2 ASM NAFIS BIOLOGY
- 32. Thanks
