Enzymes In Transcription, Translation and Replication
•Transferir como DOCX, PDF•
1 gostou•82 visualizações
I have entered here all the enzymes related to the three most important function in Molecular Biology - Replication Transcription and translation .They are concised in three simple charts. Also diagrams of 5 types of Gene that code for mRNA,rRNA,tRNA that are Transcribed by different RNA polymerase.
Denunciar
Compartilhar
Denunciar
Compartilhar
Recomendados
Recomendados
optimising HT FRET drug screening assay with ref to denaturation regimen; potentiation of soluble REL 1 production by heat shock inclusion Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...

Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...Laurence Dawkins-Hall
Mais conteúdo relacionado
Semelhante a Enzymes In Transcription, Translation and Replication
optimising HT FRET drug screening assay with ref to denaturation regimen; potentiation of soluble REL 1 production by heat shock inclusion Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...

Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...Laurence Dawkins-Hall
Semelhante a Enzymes In Transcription, Translation and Replication (20)
Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...

Labtalk 101210 optimising fret assay_den_optimising r_rel 1 production_heat s...
MOLECULAR GENETICS : PROKARYOTIC TRANSCRIPTION OR RNA SYNTHESIS BY DNA DEPEN...

MOLECULAR GENETICS : PROKARYOTIC TRANSCRIPTION OR RNA SYNTHESIS BY DNA DEPEN...
Último
Último (20)
Pests of mustard_Identification_Management_Dr.UPR.pdf

Pests of mustard_Identification_Management_Dr.UPR.pdf
High Profile 🔝 8250077686 📞 Call Girls Service in GTB Nagar🍑

High Profile 🔝 8250077686 📞 Call Girls Service in GTB Nagar🍑
Call Girls Alandi Call Me 7737669865 Budget Friendly No Advance Booking

Call Girls Alandi Call Me 7737669865 Budget Friendly No Advance Booking
Module for Grade 9 for Asynchronous/Distance learning

Module for Grade 9 for Asynchronous/Distance learning
Feature-aligned N-BEATS with Sinkhorn divergence (ICLR '24)

Feature-aligned N-BEATS with Sinkhorn divergence (ICLR '24)
9654467111 Call Girls In Raj Nagar Delhi Short 1500 Night 6000

9654467111 Call Girls In Raj Nagar Delhi Short 1500 Night 6000
High Class Escorts in Hyderabad ₹7.5k Pick Up & Drop With Cash Payment 969456...

High Class Escorts in Hyderabad ₹7.5k Pick Up & Drop With Cash Payment 969456...
dkNET Webinar "Texera: A Scalable Cloud Computing Platform for Sharing Data a...

dkNET Webinar "Texera: A Scalable Cloud Computing Platform for Sharing Data a...
Enzymes In Transcription, Translation and Replication
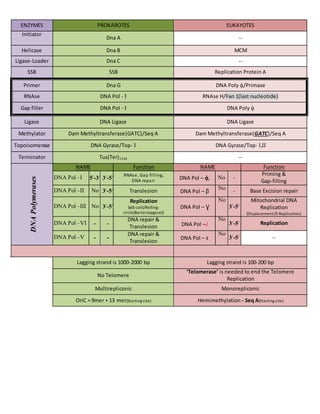
- 1. ENZYMES PROKAROTES EUKAYOTES Initiator Dna A -- Helicase Dna B MCM Ligase-Loader Dna C -- SSB SSB Replication Protein A Primer Dna G DNA Poly ᾀ/Primase RNAse DNA Pol - I RNAse H/Fan 1(last nucleotide) Gap filler DNA Pol - I DNA Poly ᾀ Ligase DNA Ligase DNA Ligase Methylator Dam Methyltransferase(GATC)/Seq A Dam Methyltransferase(GATC)/Seq A Topoisomerase DNA Gyrase/Top- I DNA Gyrase/Top- I,II Terminator Tus(Ter)1234 -- DNAPolymerases NAME Function NAME Function DNA Pol –I 5/ -3/ 3/ -5/ RNAse, Gap filling, DNA repair DNA Pol – ᾀ, No - Priming & Gap-filling DNA Pol –II No 3/ -5/ Translesion DNA Pol – β No - Base Excision repair DNA Pol –III No 3/ -5/ Replication {ɵ(E-coli)/Rolling- circle(Bacteriopgase)} DNA Pol – Ɣ No 3/ -5/ Mitochondrial DNA Replication (Displacement/D Replication) DNA Pol –VI - - DNA repair & Translesion DNA Pol –d No 3/ -5/ Replication DNA Pol –V - - DNA repair & Translesion DNA Pol – ɛ No 3/ -5/ -- Lagging strand is 1000-2000 bp Lagging strand is 100-200 bp No Telomere ‘Telomerase’ is needed to end the Telomere Replication Multirepliconic Monorepliconic OriC = 9mer + 13 mer(Starting site) Hemimethylation - Seq A(Starting site)
- 2. ENZYMES PROKARYOTES EUKARYOTES RNAPolymerases Sub Units Function Types Product Transcription Factor Function α Assembly of core enzyme, Recognition of promoter (up element) RNAPol-I 5.8s, 18s & 28s rRNA UBF(Upstream Binding factor) Binds Upstream Control Element β Catalytic centre SLI(Selectivity Factor 1) Stabilizes UBF binding β/ Catalytic centre RNAPol-II mRNA most of the sn RNA LINE miRNA TFII A Inhibits the binding of TFII D inhibitor w Assembly of RNA polymerase TFII B Additional factor helps retain TFIIA+TFIID σ Recognition of promoter(Core promoter) & initiation TFII C Not known properly TFII D (TBP + TAF) TBP(TATA binding protein) Recognize TATA box TAF(TBP associated factor) Recognition of Core promoter & Regulation of TBP TFII E Recruite TFII H TFII F Recruit RNA pol II TFII H Helicase that create “Open complex” & Phosphorylation of the C- terminal domain of largest subunit of RNA pol II TFII i Additional factor for initiationTFII J RNAPol-III tRNA 5s rRNA U6 snRNA H1 RNA, MRP RNA 7S L scRNA, 7S K scRNA SINEtranscripts TFIII A Binds with C box TFIII B Binds with A and B box TBP BRF B// TFIII C Give Direction to The TFIIIB Rho(P) Works for termination in ‘Rho dependent termination’ process --
- 3. -35 box -10 box +1 Starting point -180 -100 -45 +1 +20 Table 2 Eukaryotic rRNA gene(RNA Pol I) GC box CAATbox BRE TATA +1 MTE DPE Intr Table 3 Eukaryotic mRNA gene(RNA Pol II) +1 +50 +70 +80 +90 Table 3 Eukaryotic 5s rRNA gene(RNAPol III) Box A Box B +1 +10 +30 +50 +70 Table 3 Eukaryotic tRNA gene(RNA Pol III) DSE PSE TATA +60 +30 +1 Table 3 Eukaryotic U6 snRNA gene(RNA Pol III) Table 1 Prokaryotic Gene UCE Core Element GC CAAT BRE TATA MTE DPE Box A Box C
- 4. PROKARYOTES EUKARYOTES FUNCTION Initiation eIF3, eIF4c eIF6, eIF4B eIF4F Bind to ribosome subunit IF 1 , IF 3 Bind to mRNA Initiator tRNA delivery IF 2 eIF2B eIF2 eIF5 Displacement of other factor Elongation eEF1a Aminoacyl tRNA delivery EF-Tu EF-Ts eEFβƳ Recycling of EF-Tu/eEF1a EF-g eEF2 Translocation Termination eRF Polypeptide Chain release RF1, RF2,RF3 Ribosome Binding site(RBS) Scaning of 5/ methylated cap ISER(Poliovirus) Cap Snitching(Influenza) Shine-Dalgarno sequence KOzak Sequence AGGAGGU G/ACCAUGG