IB Chemistry on Titration Curves between Acids and Bases
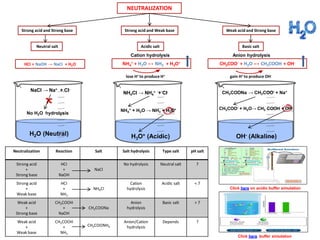
- 1. NEUTRALIZATION Neutral salt Strong acid and Strong base Strong acid and Weak base Weak acid and Strong base Acidic salt Basic salt NH4 + + H2O ↔ NH3 + H3O+ CH3COO- + H2O ↔ CH3COOH + OH- lose H+ to produce H+ gain H+ to produce OH- NH4 + + H2O → NH3 + H3O+ NH4CI → NH4 + + CI- H3O+ (Acidic) Cation hydrolysis Anion hydrolysis CH3COONa → CH3COO- + Na+ CH3COO- + H2O→ CH3 COOH + OH- OH- (Alkaline) NaCI → Na+ + CI- No H2O hydrolysis H2O (Neutral) HCI + NaOH → NaCI + H2O Neutralization Reaction Salt Salt hydrolysis Type salt pH salt Strong acid + Strong base HCI + NaOH NaCI No hydrolysis Neutral salt 7 Strong acid + Weak base HCI + NH3 NH4CI Cation hydrolysis Acidic salt < 7 Weak acid + Strong base CH3COOH + NaOH CH3COONa Anion hydrolysis Basic salt > 7 Weak acid + Weak base CH3COOH + NH3 CH3COONH4 Anion/Cation hydrolysis Depends ? Click here on acidic buffer simulation Click here buffer simulation
- 2. CH3COO- + H2O → CH3 COOH + OH- Salt Hydrolysis Neutralization Reaction Salt Salt hydrolysis Type salt pH salt Strong acid + Strong base HCI + NaOH NaCI No hydrolysis Neutral salt 7 Strong acid + Weak base HCI + NH3 NH4CI Cation hydrolysis Acidic salt < 7 Weak acid + Strong base CH3COOH + NaOH CH3COONa Anion hydrolysis Basic salt > 7 Weak acid + Weak base CH3COOH + NH3 CH3COONH4 Anion/Cation hydrolysis Depends ? Weak acid and Weak base CH3COOH + NH3 → CH3COONH4 Acidicity depend on Ka and Kb Ka > Kb – Acidic – H+ ions produced Kb < Ka – Basic – OH- ions produced Ka = Kb – Neutral – hydrolyzed same extent. CH3COONH4 → CH3COO- + NH4 + NH4 + + H2O → NH3 + H3O+ salt anion cation OH- - Basic H3O+ - AcidicKb Ka Ka = Kb NEUTRAL NH3 + HF → NH4F salt NH4F → NH4 + + F- NH4 + + H2O → NH3 + H3O+ F- + H2O → HF + OH- cation anion Ka H3O+ - Acidic Kb OH- - Basic Acidicity depend on Ka and Kb Ka > Kb – Acidic – H+ ions produced Kb < Ka – Basic – OH- ions produced Ka = Kb – Neutral – hydrolyzed same extent. Kb > Ka BASIC Weak acid + Weak base
- 3. Titration bet strong acid with strong base HCI + NaOH → NaCI + H2O TitrationcurvesStrong Acid with Strong Base Click here titration simulation NaOH M = 0.1M V = 0 ml HCI M = 0.1M V = 25ml 7 HCI + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 2.7 11.3 • Rapid jump in pH (2.7 – 11.3) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, NaCI - neutral 1 HCI M = 0.1M V = 25ml NaOH M = 0.1M V = 25ml HCI left → 25 ml, 0.1M Conc H+ = 0.1M HCI M = 0.1M V = 1ml left NaOH M = 0.1M V = 24 ml add HCI left → 1ml, 0.1M Mole H+ = (0.1 x 1)/1000 = 0.0001mol Conc H+ = Mole/Vol = 0.0001/0.049 = 0.002M NaOH M = 0.1M V = 25ml add HCI M = 0.1M V = 0ml left Neutral Salt, NaCI Conc H+ = 1 x 10-7M (Dissociation of water) NaOH M = 0.1M V = 26ml add NaOH left → 1ml left, 0.1M Moles OH-= (0.1 x 1)/1000 = 0.0001mol Conc OH- = Moles/Vol = 0.0001/0.051 = 0.002M NaOH V = 1ml left Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml 11.3 2.7 Neutralization Mole ratio 1: 1 Equivalent point pH 7 Dilution factor! 1 ]1.0lg[ ]lg[ pH pH HpH 7.2 ]002.0lg[ ]lg[ pH pH HpH 7 ]101lg[ ]lg[ 7 pH pH HpH 3.117.214 7.2 )002.0lg( )lg( pH pOH pOH OHpOH 7
- 4. Titration bet strong acid with weak base HCI + NH4OH → NH4CI + H2O TitrationcurvesStrong Acid with Weak Base NH4OH M = 0.1M V = 0 ml HCI M = 0.1M V = 25ml 5.3 HCI + NH4OH → NH4CI + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 2.7 7.8 • Rapid jump in pH (2.7 – 7.8) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 5.3 • Acidic salt, NH4CI – pH = 5.3 1 HCI M = 0.1M V = 25ml NH4OH M = 0.1M V = 25ml HCI left → 25 ml, 0.1M Conc H+ = 0.1M HCI M = 0.1M V = 1ml left NH4OH M = 0.1M V = 24 ml add HCI left → 1ml, 0.1M Mole H+ = (0.1 x 1)/1000 = 0.0001mol Conc H+ = Mole/Vol = 0.0001/0.049 = 0.002M NH4OH M = 0.1M V = 25ml add HCI M = 0.1M V = 0ml left Acidic Salt, NH4CI NH4 + hydrolysis to produce H+ pH = 5.3 NH4OH M = 0.1M V = 26ml add NH4OH left → 1ml left, 0.1M Moles OH-= (0.1 x 1)/1000 = 0.0001mol Conc OH- = Moles/Vol = 0.0001/0.051 = 0.002M Conc NH4CI = Moles/Vol = 2.5 x 10-3/0.051 = 0.05M NH4OH V = 1ml left Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml 7.8 2.7 Neutralization Click here titration simulation pOH = pKb -lg (base) (salt) pOH = 4.74 – lg (0.002) (0.05) pOH = 6.13 pH + pOH = 14 pH = 7.8 pH buffer – salt and weak base pH buffer region salt and weak base 1 ]1.0lg[ ]lg[ pH pH HpH 7.2 ]002.0lg[ ]lg[ pH pH HpH 5.3 Mole ratio 1: 1
- 5. Titration between weak acid with strong base CH3COOH + NaOH → CH3COONa + H2O TitrationcurvesWeak Acid with Strong Base NaOH M = 0.1M V = 0 ml NaOH M = 0.1M V = 25ml 9 CH3COOH + NaOH → CH3COONa + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 11.3 6.11 • Rapid jump in pH (6.11 – 11.3 ) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 9 • Basic salt, CH3COONa = pH 9 2.87 CH3COOH M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml CH3COOH left → 25 ml, 0.1M CH3COOH ↔ (CH3COO- )(H+) Ka = (CH3COO-) (H+) CH3COOH (H+) = √Ka x CH3COOH (H+) = 1.34 x 10-3 CH3COOH M = 0.1M V = 1ml left NaOH M = 0.1M V = 24 ml add NaOH M = 0.1M V = 25ml add CH3COOH M = 0.1M V = 0ml left Basic Salt (CH3COONa) CH3COO- hydrolysis produce OH- pH = 9 NaOH M = 0.1M V = 26ml add NaOH V = 1ml left Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml Click here titration simulation CH3COOH left → 1ml, 0.1M Mole CH3COOH = (MV)/1000 = (1 x 0.1)/1000 = 0.0001mol Conc CH3COOH = Mole/Vol = 0.0001/0.049 = 2.04 x 10-3 Conc CH3COONa = Mole/Vol = 2.4 x 10-3/0.049 = 0.048M Ka = 1.8 x 10-5 NaOH left → 1ml left, 0.1M Moles OH-= (0.1 x 1)/1000 = 0.0001mol Conc OH- = Moles/Vol = 0.0001/0.051 = 0.002M 11.3 6.11 Neutralization pH buffer region weak acid and salt pH buffer – salt and weak acid pH = pKa -lg [acid] [salt] pH = 4.74 – lg [2.04 x 10-3] [0.048] pH = 4.74 + 1,37 pH = 6.11 Weak Acid 87.2 ]1034.1lg[ ]lg[ 3 pH pH HpH 9 3.117.214 7.2 )002.0lg( )lg( pH pOH pOH OHpOH Mole ratio 1: 1
- 6. TitrationcurvesWeak Acid with Weak Base NH4OH M = 0.1M V = 0 ml CH3COOH M = 0.1M V = 25ml 7 CH3COOH + NH4OH → CH3COONH4 + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 7.8 • No sharp rise in pH • pH changes gradually over a range • No inflection point 2.87 CH3COOH M = 0.1M V = 25ml NH4OH M = 0.1M V = 25ml NH4OH M = 0.1M V = 25ml add CH3COOH M = 0.1M V = 0ml left Neutral Salt CH3COONH4 pH = 7 NH4OH M = 0.1M V = 26ml add NH4OH V = 1ml left Total = 25 + 26 Vol = 51ml 7.8 6.11 Neutralization Titration between weak base with weak acid CH3COOH + NH4OH → CH3COONH4 + H2O 6.11 NH4OH M = 0.1M V = 24ml add CH3COOH M = 0.1M V = 1ml left Click here titration simulation CH3COOH left → 25 ml, 0.1M CH3COOH ↔ (CH3COO- )(H+) Ka = (CH3COO-) (H+) CH3COOH (H+) = √Ka x CH3COOH CH3COOH left → 1ml, 0.1M Mole CH3COOH = (MV)/1000 = (1 x 0.1)/1000 = 0.0001mol Conc CH3COOH = Mole/Vol = 0.0001/0.049 = 2.04 x 10-3 Conc CH3COONH4 = Moles/Vol = 2.4 x 10-3/0.049 = 0.048M pH = pKa -lg [acid] [salt] pH = 4.74 – lg [2.04 x 10-3] [0.048] pH = 4.74 + 1,37 pH = 6.11 pH buffer – weak acid and salt pH buffer region weak acid and salt NH4OH left → 1ml, 0.1M pH = 7.8 87.2 ]1034.1lg[ ]lg[ 3 pH pH HpH Click here for notes Mole ratio 1: 1
- 7. Titration bet strong acid with strong base HCI + NaOH → NaCI + H2O Titration bet weak acid with strong base CH3COOH + NaOH → CH3COONa + H2O Titrationcurves Acid with Base Titration bet strong acid with weak base HCI + NH4OH → NH4CI + H2O 11.3 2.7 Titration bet weak acid with weak base CH3COOH + NH4OH → CH3COONH4 + H2O NaOH M = 0.1M V = 25ml 6.11 11.3 NaOH M = 0.1M V = 25ml 2.87 1 Vs •Start at pH = 1 → End at 11.3 • Rapid change at equivalence pt • Rapid jump in pH (2.7 – 11.3) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, NaCI - neutral •Start at pH = 2.87 → End at 11.3 • Rapid change at equivalence pt • Rapid jump in pH (6.11– 11.3) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 9 • Basic salt, CH3COONa - basic 9 7 Vs •Start at pH = 1 → End at 7.8 • Rapid change at equivalence pt • Rapid jump in pH (2.7 – 7.8) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 5.3 • Acidic salt, NH4CI - acidic 1 2.7 5.3 7.8 2.87 NH4OH M = 0.1M V = 25 ml NH4OH M = 0.1M V = 25 ml •Start at pH = 2.87 → End at 7.8 • pH changes gradually over a range • No sharp rise in pH • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, CH3COONH4 - neutral 6.11 7 7.8 pH buffer region salt and weak base pH buffer region salt and weak acid Buffer region form • Slow gradual increase pH due to buffering effect Buffer region form • Slow gradual increase pH due to buffering effect HCI M = 0.1M V = 25ml HCI M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml
- 8. Strong acid vs Strong base Titration Acid Base Strong acid vs Weak base Weak acid vs Strong base Weak acid vs Weak base Acid Base Indicator 2.7 11.3 7.8 2.7 11.3 6.11 7.8 6.11 HCI M = 0.1M V = 25ml Dilution Factor Water Adding 20 ml water What is conc H+ and pH? 3 105.2.. 025.01.0.. .. HMole HMole VMHMole 1 ]1.0lg[ ]lg[ pH pH HpH 055.0...... 045.0 105.2 .. 3 HConc Volume Mole HConc Before adding Water After adding Water Vol/Conc change 25.1 ]055.0lg[ ]lg[ pH pH HpH pH drop due dilution Factor Adding 20 ml base NaOH What is conc H+ and pH? NaOH HCI M = 0.1M V = 25ml Total vol = 25 + 20 = 45ml Dilution/Neutralization Factor Before adding NaOH 3 105.2.. 025.01.0.. .. HMole HMole VMHMole 1 ]1.0lg[ ]lg[ pH pH HpH Mole change After adding NaOH 3 105.0... leftHMole 011.0. 045.0 105.0 .. 3 HConc Volume Mole HConc 95.1 ]011.0lg[ ]lg[ pH pH HpH Total vol = 25 + 20 = 45ml pH drop due dilution/Neutralization Factor Dilution Factor during Titration Why adding water and base causes pH to increase?
- 9. TitrationcurvesStrong Acid with Strong Base HCI M = 0.1M V = 0 ml HCI M = 0.1M V = 25ml 7 HCI + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 2.7 11.3 • Rapid drop in pH (11.3–2.7 ) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, NaCI - neutral NaOH M = 0.1M V = 25ml NaOH M = 0.1M V = 25ml NaOH left → 25 ml, 0.1M Conc OH- = 0.1M NaOH M = 0.1M V = 1ml left HCI M = 0.1M V = 24 ml add NaOH left → 1ml, 0.1M Mole OH- = (0.1 x 1)/1000 = 0.0001mol Conc OH- = Mole/Vol = 0.0001/0.049 = 0.002M HCI M = 0.1M V = 25ml add NaOH M = 0.1M V = 0 ml left Neutral Salt, NaCI Conc H+ = 1 x 10-7M (dissociation of water) HCI M = 0.1M V = 26ml add HCI left → 1ml left, 0.1M Moles H+ = (0.1 x 1)/1000 = 0.0001mol Conc H+ = Moles/Vol = 0.0001/0.051 = 0.002M HCI V = 1ml left Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml Click here titration simulation 13 11.3 2.7 Neutralization Titration bet strong base with strong acid HCI + NaOH → NaCI + H2O 13114 1 )1.0lg( )lg( pH pOH pOH OHpOH 3.117.214 7.2 )002.0lg( )lg( pH pOH pOH OHpOH 7 ]101lg[ ]lg[ 7 pH pH HpH 7.2 ]002.0lg[ ]lg[ pH pH HpH Mole ratio 1: 1
- 10. TitrationcurvesWeak Acid with Strong Base CH3COOH M = 0.1M V = 0 ml NaOH M = 0.1M V = 25ml 9 CH3COOH + NaOH → CH3COONa + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 11.3 6.13 • Rapid drop in pH ( 11.3 - 6.13) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 9 • Basic salt, CH3COONa = pH 9 13 NaOH M = 0.1M V = 25.0ml CH3COOH M = 0.1M V = 25ml NaOH M = 0.1M V = 1ml left CH3COOH M = 0.1M V = 24 ml add CH3COOH M = 0.1M V = 25ml add NaOH M = 0.1M V = 0ml left Basic Salt (CH3COONa) CH3COO- hydrolysis to produce OH- pH = 9 CH3COOH M = 0.1M V = 26ml add CH3COOH V = 1ml left Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml Click here titration simulation NaOH left → 25 ml, 0.1M Conc OH- = 0.1M NaOH left → 1ml, 0.1M Moles OH- = (0.1 x 1)/1000 = 0.0001mol Conc OH- = Mole/Vol = 0.0001/0.049 = 0.002M CH3COOH left → 1ml, 0.1M Mole CH3COOH = (MV)/1000 = (1 x 0.1)/1000 = 0.0001mol Conc CH3COOH = Mole/Vol = 0.0001/0.051 = 1.96 x 10-3 Conc CH3COONa = Mole/Vol = 2.5 x 10-3/0.051 = 0.049M 11.3 6.13 Neutralization pH buffer region weak acid + salt Titration bet strong base with weak acid CH3COOH + NaOH → CH3COONa + H2O pH = pKa -lg [acid] [salt] pH = 4.74 – lg [1.96 x 10-3] [0.049] pH = 4.74 + 1,39 pH = 6.13 pH buffer – salt and weak acid 13114 1 )1.0lg( )lg( pH pOH pOH OHpOH 3.117.214 7.2 )002.0lg( )lg( pH pOH pOH OHpOH Mole ratio 1: 1
- 11. TitrationcurvesStrong Acid with Weak Base HCI M = 0.1M V = 0 ml HCI M = 0.1M V = 25ml 5.3 HCI + NH4OH → NH4CI + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 7.8 2.7 • Rapid drop in pH (7.8 – 2.7) • Rapid change at equivalence pt • Equivalence pt → amt acid = amt base • pH at equivalence pt = 5.3 • Acidic salt, NH4CI – pH = 5.3 11.1 NH4OH M = 0.1M V = 25ml NH4OH M = 0.1M V = 1ml left HCI M = 0.1M V = 24 ml add HCI M = 0.1M V = 25ml add Acidic Salt, NH4CI NH4 + hydrolysis to produce H+ pH = 5.3 HCI M = 0.1M V = 26ml add NH4OH left → 1ml left, 0.1M Mole NH4OH = (0.1 x 1)/1000 = 0.0001 mol Conc NH4OH = Mole/Vol = 0.0001/0.049 = 0.002M Conc NH4CI = Mole/Vol = 2.4 x 10-3/0.049 = 0.048 Total = 24 + 25 Vol = 49ml Total = 25 + 26 Vol = 51ml 7.8 2.7 Neutralization pOH = pKb -lg (base) (salt) pOH = 4.74 – lg (0.002) = 6.12 (0.048) pH + pOH = 14 pH = 7.8 pH buffer – Weak base + salt Click here titration simulation NH4OH left → 25 ml, 0.1M NH4OH ↔ NH4 + + OH- Kb = (NH4 + ) (OH- ) (NH4OH) 1.8 x 10-5 = (OH- )2 0.1 OH- = √0.1 x 1.8 x 10-5 OH- = 1.34 x 10-3 pH buffer region weak base + salt HCI left → 1ml left, 0.1M Mole H+ = (0.1 x 1)/1000 = 0.0001mol Conc H+ = Mole/Vol = 0.0001/0.051 = 0.002M Titration bet weak base with strong acid HCI + NH4OH → NH4CI + H2O NH4OH M = 0.1M V = 0ml left 1.1187.214 87.2 )1034.1lg( )lg( 3 pH pOH pOH OHpOH NH4OH M = 0.1M V = 25ml HCI V = 1ml left 7.2 ]002.0lg[ ]lg[ pH pH HpH Mole ratio 1: 1
- 12. TitrationcurvesWeak Acid with Weak Base CH3COOH M = 0.1M V = 0 ml CH3COOH M = 0.1M V = 25ml7 CH3COOH + NH4OH → CH3COONH4 + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml 6.11 • No sharp drop in pH • pH changes gradually over a range • no inflection point 11.1 NH4OH M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml add Neutral Salt CH3COONH4 pH = 7 CH3COOH M = 0.1M V = 26ml add Total = 25 + 26 Vol = 51ml 7.8 6.11 Neutralization Click here titration simulation NH4OH left → 25 ml, 0.1M Conc NH4OH = 0.1M NH4OH ↔ NH4 + + OH- Kb = (NH4 + ) (OH- ) (NH4OH) 1.8 x 10-5 = (OH- )2 0.1 OH- = √0.1 x 1.8 x 10-5 OH- = 1.34 x 10-3 Titration bet weak base with weak acid CH3COOH + NH4OH → CH3COONH4 + H2O CH3COOH left → 1ml, 0.1M pH = 6.11 7.8 CH3COOH M = 0.1M V = 24ml add NH4OH left → 1ml left, 0.1M Mole NH4OH = (0.1 x 1)/1000 = 0.0001mol Conc NH4OH = Mole/Vol = 0.0001/0.049 = 0.002M Conc NH4CI = Mole/Vol = 2.4 x 10-3/0.049 = 0.048M pOH = pKb -lg (base) (salt) pOH = 4.74 – lg (0.002) (0.048) pOH = 6.12 pH + pOH = 14 pH = 7.8 pH buffer – weak base + salt pH buffer region weak base + salt NH4OH M = 0.1M V = 25ml NH4OH M = 0.1M V = 1ml left NH4OH M = 0.1M V = 0ml left 1.1187.214 87.2 )1034.1lg( )lg( 3 pH pOH pOH OHpOH CH3COOH V = 1ml left Mole ratio 1: 1
- 13. Titration bet strong base with strong acid HCI + NaOH → NaCI + H2O Titration bet strong base with weak acid CH3COOH + NaOH → CH3COONa + H2O Titrationcurves Acid with Base Titration bet weak base with strong acid HCI + NH4OH → NH4CI + H2O 11.3 2.7 Titration bet weak base with weak acid CH3COOH + NH4OH → CH3COONH4 + H2O NaOH M = 0.1M V = 25ml HCI M = 0.1M V = 25ml 6.13 11.3 NaOH M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml 13 Vs •Start at pH = 1 3 → End at 2.7 • Rapid change at equivalence pt • Rapid drop in pH (11.3 – 2.7) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, NaCI - neutral •Start at pH = 13 → End at 6.13 • Rapid change at equivalence pt • Rapid drop in pH (11.3 – 6.13) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 9 • Basic salt, CH3COONa - basic 9 7 Vs •Start at pH = 11.1 → End at 2.7 • Rapid change at equivalence pt • Rapid drop in pH (7.8 – 2.7) • Equivalence pt → amt acid = amt base • pH at equivalence pt = 5.3 • Acidic salt, NH4CI - acidic 11.1 2.7 5.3 7.8 11.1 NH4OH M = 0.1M V = 25 ml HCI M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml •Start at pH = 11.1 → End at 6.11 • pH changes gradually over a range • No sharp drop in pH • Equivalence pt → amt acid = amt base • pH at equivalence pt = 7 • Neutral salt, CH3COONH4 - neutral 6.11 7 7.8 13 Buffer region form • Slow gradual drop pH due to buffering effect Buffer region form • Slow gradual drop pH due to buffering effect pH buffer region weak acid + salt pH buffer region weak base + salt NH4OH M = 0.1M V = 25 ml
- 14. Acidic Buffer Region CH3COOH (acid)/CH3COO- (salt) Titration bet weak acid + strong base CH3COOH + NaOH → CH3COONa + H2O Click here buffer simulation CH3COOH + NaOH → CH3COONa + H2O Initial 0.0025 mol 0.00125mol added 0 Change (0.0025-0.00125)mol 0 mol 0.00125 mol form Final 0.00125mol left 0 mol 0.00125 mol form At half equivalencepoint : • Amt acid = Amt salt : ( 0.00125 = 0.00125) • pH = pKa -lg [acid] → pH = pKa → 4.74 = 4.74 [salt] Buffer at pH = 4.74 form when half amt of acid neutralize by base or at half equivalencept when amt acid = amt salt Prepare Acidic Buffer pH = 4.74 • Choose pKa acid closest pH 4.74 • pKa = 4.74 (ethanoic acid) chosen • pH = pKa -lg [acid] [salt] • 4.74 = 4.74 – lg [acid] [salt] • [acid] = 1.00 (amt acid = amt salt) [salt] Buffer region at half equivalence pt Amt acid = Amt salt Weak acid 25ml, 0.1M (0.0025 mol) Titration bet weak acid with strong base CH3COOH + NaOH → CH3COONa + H2O Buffer region at half equivalence point Amt acid = Amt salt At equivalence point (Neutralization) Amt acid = Amt base CH3COOH + NaOH → CH3COONa + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml At half equivalence point : • Vol base = 12.5ml • pH = pKa • Most effective buffering capacity At equivalence point: • vol base = 25ml • Amt acid = amt base • Neutralization • Salt and water NaOH M = 0.1M V = 25 ml add NaOH M = 0.1M V = 12.5 ml add CH3COOH M = 0.1M V = 12.5ml left CH3COOH M = 0.1M V = 0ml left Strong base 12.5ml, 0.1M add (0.00125 mol) pH buffer region weak acid + salt Titration curve use to find pKa or Ka for weak acid pH = pKa Completely neutralize Half neutralize Mole ratio 1: 1
- 15. NH4OH + HCI → NH4CI + H2O Initial 0.0025 mol 0.00125 mol add 0. Change (0.0025-0.00125)mol 0 0.00125 mol form Final 0.00125 mol left 0 0.00125 mol form At half equivalencepoint : • Amt base = Amt salt : (0.00125 = 0.00125) • pOH = pKb - lg [base] → pOH = pKb → 4.74 = 4.74 [salt] Buffer at pOH = 4.74 form when half amt of base neutralise by acid or at half equivalence point when amt base = amt salt Prepare Buffer pH = 9.26 /pOH = 4.74 • Choose pKb base closest to pOH = 4.74 • pKb = 4.74 (NH3) chosen • pOH = pKb -lg [base] [salt] • 4.74 = 4.74 – lg [base] [salt] • [base] = 1.00 (amt base = amt salt) [salt] NH4OH + HCI → NH4CI + H2O M = 0.1M M = 0.1M V = 25ml V = 25ml Titration bet weak base + strong acid NH4OH + HCI → NH4CI + H2O Click here buffer simulation Buffer region at half equivalence pt Amt acid = Amt salt Weak base 25ml, 0.1M (0.0025 mol) Titration bet weak base with strong acid NH4OH + HCI → NH4CI + H2O Buffer region at half equivalence point Amt acid = Amt salt At equivalence point (Neutralization) Amt acid = Amt base At half equivalence point : • Vol acid = 12.5ml • pH = pKb • Most effective buffering capacity At equivalence point: • vol acid = 25ml • Amt acid = amt base • Neutralization • Salt and water HCI M = 0.1M V = 25 ml add HCI M = 0.1M V = 12.5 ml add NH4OH M = 0.1M V = 12.5ml left NH4OH M = 0.1M V = 0ml left Strong acid 12.5ml, 0.1M add (0.00125 mol) pH buffer region weak base + salt Titration curve use to find pKb or Kb for weak base pH = pKb Basic Buffer Region NH3(base)/NH4CI(salt) Completely neutralize Half neutralize Mole ratio 1: 1
- 16. Click here view buffering Concept Map Buffer pH Proton availability Stable Buffer solution Weak acid ↔ Conjugate base ][ ][ lg salt acid pKpH a pH = -lg[H+] made up of HA ↔ H+ + A- Weak base ↔ Conjugate acid or Buffering capacity highest Buffer formula pH = pKa 1 ][ ][ Salt Acid B + H2O ↔ BH+ + OH- or Ratio of acid salt Dilution Add water pH buffer pH will not change Temperature affect pH pH change Strong acid Strong base Titration Acid Base Strong acid Weak base Weak acid Strong base Weak acid Weak base Neutralization Titration curve Base Acid Indicator 2.7 11.3 7.8 2.7 11.3 6.11 7.8 6.11 11.3 2.7 7.8 2.7 11.3 6.11 7.8 6.11 Indicator Acid Base Adding base to acid Adding acid to base
- 17. Concept Map Strong acid Strong base Titration Acid Base Strong acid Weak base Weak acid Strong base Weak acid Weak base Neutralization Titration curve Base Acid Indicator 2.7 11.3 7.8 2.7 11.3 6.11 7.8 6.11 Adding base to acid End point pH range at equivalent pt Equivalent pt Stoichiometricpt Point of inflection Titration bet weak acid with strong base CH3COOH + NaOH → CH3COONa + H2O pH buffer region weak acid and salt 6.11 11.3 9 • Amt acid = Amt base • Vol at equivalencept = 25ml 25ml12.5ml Half Equivalent pt • Amt acid = Amt salt • Vol is = 12.5ml • Buffer region form Indicator change colour Titration bet strong acid with weak base HCI + NH4OH → NH4CI + H2O 25ml Equivalent pt Stoichiometric pt • Amt acid = Amt base • Vol at equivalencept = 25ml pH = pKa 6.3 2.7 7.8 pH buffer region weak base and salt Buffer region 50ml pOH = pKb • Amt base = Amt salt • Vol is = 50 ml • Buffer region form
- 18. CH3COOH + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25 ml V = ? ml Titration betweenStrong Acid with Strong Base HCI + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25 ml V = 25 ml HCI M = 0.1M V = 25 ml NaOH M = 0.1M V = ? ml 25 ml, 0.1M strong acid need to neutralize, 25ml, 0.1M strong base? STRONG BASE Mole ratio – 1: 1 Mole ratio – 1: 1 NaOH M = 0.1M V = 25 ml 2.5 x 10-3 mol NaOH 2.5 x 10-3 H+ ion 2.5 x 10-3 mol HCI Strong acid/base dissociate completely Titration between Weak Acid with Strong Base What vol of 0.1M strong base need to neutralize, 25ml, 0.1M weak acid? CH3COOH M = 0.1M V = 25 ml Will 2.5 x 10-3 mol weak acid dissociate completely? Answer – Yes 25ml base needed Regardless whether strong or weak acid/base • Stoichiometric mole ratio is follow for neutralization • 2.5 x 10-3 mol weak acid neutralize 2.5 x 10-3 mol strong base CH3COOH + H2O ↔ H3O+ + CH3COO- H3O+ + OH- ↔ 2H2O CH3COOH + OH- → CH3COO- + H2O Le Chatelier principle ↓ Addition OH- remove H+ ↓ Conc H+ reduced ↓ Shift equilibrium right ↓ Weak acid, CH3COOH dissociate completely ↓ Produce 2.5 x 10-3 mol H+ ↓ Volume Strong base = 25 ml WEAK ACID CH3COOH STRONG BASE NaOH CH3COOH ↔ CH3COO- + H+ OH- CH3COOH + OH- → CH3COO- + H2O 5 108.1 aK 14 100.1 wK 9 145 108.1 100.1108.1 rxn rxn wbrxn K K KKK ANSWER Effect on Kc Inverse rxn Add 2 rxn wK 1 wb KK + Krxn HIGH •Shift to right •Weak acid dissociate completely STRONG ACID
- 19. HCI + NH4OH → NH4CI + H2O M = 0.1M M = 0.1M V = ? ml V = 25 ml Titration betweenStrong Acid with Strong Base HCI + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25 ml V = 25 ml HCI M = 0.1M V = 25 ml NH4OH M = 0.1M V = 25 ml 25 ml, 0.1M strong acid need to neutralize, 25ml, 0.1M strong base? STRONG ACID STRONG BASE Mole ratio – 1: 1 Mole ratio – 1: 1 NaOH M = 0.1M V = 25 ml 2.5 x 10-3 mol NaOH 2.5 x 10-3 OH- ion 2.5 x 10-3 mol HCI Strong acid/base dissociate completely Titration between Strong Acid with Weak Base What vol of 0.1M strong acid need to neutralize, 25ml, 0.1M weak base? HCI M = 0.1M V = ? ml Will 2.5 x 10-3 mol weak base dissociate completely? Answer – Yes 25ml acid needed Regardless whether strong or weak acid/base • Stoichiometric mole ratio is follow for neutralization • 2.5 x 10-3 mol strong acid neutralize 2.5 x 10-3 mol weak base NH3 + H2O ↔ NH4 + + OH- H+ + OH- ↔ H2O NH3 + H+ → NH4 + Le Chatelier principle ↓ Addition H+ remove OH- ↓ Conc OH- reduced ↓ Shift equilibrium right ↓ Weak base, NH4OH dissociate completely ↓ Produce 2.5 x 10-3 mol OH- ↓ Volume Strong acid = 25 ml STRONG ACID HCI WEAK BASE NH4OH → NH4 + + OH- NH4OH H+ NH4OH + HCI → NH4CI + H2O 5 108.1 bK 14 100.1 wK 9 145 108.1 100.1108.1 rxn rxn wbrxn K K KKK ANSWER Effect on Kc Inverse rxn Add 2 rxn wK 1 wb KK + Krxn HIGH •Shift to right •Weak base dissociate completely NH3 molecules dissociates completely
- 20. Neutralization acid and base Strong base with Strong acid HCI + NaOH → NaCI + H2O M = 0.1M M = 0.1M V = 25 ml V = ? CH3COOH + NH4OH → CH3COONH4 + H2O M = 0.1M M = 0.1M V = 25ml V = ? CH3COOH + NaOH → CH3COONa + H2O M = 0.1M M = 0.1M V = 25ml V = ? Weak base with Strong acid Weak base with Weak acid HCI M = 0.1M V = 25ml HCI + NH4OH → NH4CI + H2O M = 0.1M M = 0.1M V = 25 ml V = ? CH3COOH M = 0.1M V = 25ml HCI M = 0.1M V = 25ml CH3COOH M = 0.1M V = 25ml Will volume used the same? Will volume used the same? Yes 25ml Yes 25ml Stoichiometric mole ratio is followed for neutralization • 1 mole weak/strong acid will neutralize 1 mole weak/strong base STRONG ACID WEAK ACID WEAK BASE STRONG ACID WEAK ACID Mole ratio – 1: 1 Mole ratio – 1: 1 Mole ratio – 1: 1 Mole ratio – 1: 1 NaOH M = 0.1M V = ? STRONG BASE STRONG BASE NaOH M = 0.1M V = ? Strong base with Weak acid NH4OH M = 0.1M V = ? WEAK BASE NH4OH M = 0.1M V = ?
- 21. IB Buffer Calculation Find pH buffer , titration by adding 18ml, 0.10M HCI to 32ml, 0.10M NH3 pKb = 4.751 Titration bet strong acid with weak base NH3 + HCI → NH4CI + H2O Buffer region Weak base + salt NH3 + HCI → NH4CI + H2O Initial 3.2 x 10-3 mol 1.8 x 10-3 mol add 0 0 Change (3.2 – 1.8) x 10-3 0 1.8 x 10-3 mol form Final 1.4 x 10-3 0 1.8 x 10-3 mol form Change mole to Conc → Mole ÷ Total vol Conc (1.4 x 10-3)/ 0.05 (1.8 x 10-3)/0.05 Conc 2.8 x 10-2 M 3.6 x 10-2 M (base) (salt) • pOH = pKb - lg [base] [salt] • pOH = 4.75 – lg [2.8 x 10-2]/[3.6 x 10-2] • pOH = 4.86 pH + pOH = 14 pH = 9.14 Click here buffer simulation Strong acid 18ml, 0.1M HCI add Weak base 32ml, 0.1M NH3 Total vol = 50 ml or 0.05 dm3 NH3 + H2O ↔ NH4 + + OH- Kb = (NH4 + ) (OH- ) (NH3) 1.77 x 10-5 = 3.6 x 10-2 x OH- 2.8 x 10-2 OH- = 2.8 x 10-2 x 1.77 x 10-5 3.6 x 10-2 OH- = 1.37 x 10-5 pOH = -lg[OH-] pOH = -lg 1.37 x 10-5 pOH = 4.86 pH + pOH = 14 pH = 9.14 1st method (formula) 2nd method (Kb) pH calculation molVMmole 3 108.1 1000 1.00.18 molVMmole 3 102.3 1000 1.032
- 22. 2 Find pH when 50ml 0.1M NaOH add to 100ml, 0.1M CH3COOH Ka CH3COOH = 1.8 x 10-5M, pKa = 4.74 Titration bet strong base with weak acid NaOH + CH3COOH → CH3COONa + H2O Click here buffer simulation Buffer region at half equivalence point Amt base = Amt salt Buffer Calculation 1st method (formula) 2nd method (Ka) NaOH + CH3COOH → CH3COONa H2O Initial 5 x 10-3 mol add 10 x 10-3 mol 0 0 Change 0 (10-5) x 10-3 5 x 10-3 mol form Final 0 5 x 10-3 5 x 10-3 mol form Change mole to Conc → Mole ÷ Total Vol Conc (5 x 10-3)/0.15 (5 x 10-3)/0.15 Conc 3.3 x 10-2 M 3.3 x 10-2 M (acid) (salt) • pH = pKa - lg [acid] [salt] • pH = 4.74 – lg [3.3 x 10-2]/[3.3 x 10-2] • pH = 4.74 Total vol = 150 ml or 0.15 dm3 CH3COOH ↔ CH3COO- +H+ Ka = (CH3COO- )(H+ ) (CH3COOH) 1.8 x 10-5 = 3.3 x 10-2 x (H+ ) 3.3 x 10-2 H+ = 1.8 x 10-5 pH = -lg [H+ ] pH = -lg(1.8 x 10-5 ) pH = 4.74 pH calculation molVMmole 3 105 1000 1.050 molVMmole 3 1010 1000 1.0100 Strong base 50ml, 0.1M NaOH add Weak acid 100ml, 0.1M CH3COOH
- 23. pH Calculation 3 Find pH when 50ml 0.1M NaOH add to 100ml, 0.1M HCI Strong base 50ml, 0.1M NaOH add Strong acid 100ml, 0.1M HCI Click here buffer simulation Titration bet strong base with strong acid NaOH + HCI → NaCI + H2O pH calculation NaOH + HCI → NaCI + H2O Initial 5 x 10-3 mol added 10 x 10-3 mol 0 0 Change 0 (1 0 - 5) x 10-3 Final 0 5 x 10-3 Change mole to Conc → Mole ÷ Total Vol Conc (5 x 10-3)/ 0.15 Conc 3.3 x 10-2 M • pH = -lg[H+] • pH = -lg 3.3 x 10-2 pH = 1.48 Total vol = 150 ml or 0.15 dm3 NH4 + + H2O ↔ NH3 + H3O+ Ka = (NH3)(H3O+ ) (NH4 + ) (H3O+ )2 = Ka x NH4 + H+ = √5.56 x 10-10 x 0.10 H+ = 7.45 x 10-6 pH = -lg 7.45 x 10-6 pH = 5.13 Find pH 0.10M NH4CI in water. Kb NH3 = 1.8 x 10-5 M4 Acid dissociation constant 0.10M NH4CI Ka (NH4) x Kb(NH3) = Kw Ka = Kw /Kb Ka = 10-14/ 1.8 x 10-5 Ka = 5.56 x 10-10 Using Ka Find pH 0.50M NH3 in water. Kb NH3 = 1.8 x 10-5 M5 NH3 + H2O ↔ NH4 + + OH- Kb = (NH4 + ) (OH- ) (NH3) 1.8 x 10-5 = (OH- )2 0.50 OH- = √0.50 x 1.8 x 10-5 OH- = 3.0 x 10-3 pOH = -lg 3.0 x 10-3 pOH = 2.52 pH = 14 – 2.52 pH = 11.48 0.50M NH3 Base Dissociation constant Using Kb molVMmole 3 105 1000 1.050 molVMmole 3 1010 1000 1.0100
- 24. IB Questions 6 Table shows Ka values for acids at 298K. Write expression for Ka. Arrange acid in order of increasing strength 7 Acid CH3COOH HCN HSO4 - Ka 1.8 x 10-5 4.9 x 10-10 1.2 x 10-2 Table shows Kb values for base at 298K. Write expression for Kb. Arrange base in order of increasing strength Base C2H5NH2 N2H4 NH3 Kb 4.7 x 10-4 4.9 x 10-10 1.2 x 10-2 CH3COOH ↔ CH3COO- + H+ COOHCH HCOOCH Ka 3 3 HCN + H2O ↔ CN- + H3O+ HCN OHCN Ka 3 HSO4 - + H2O ↔ SO4 2- + H3O+ 4 3 2 4 HSO OHSO Ka Ka – Highest – Strongest acidKa – Lowest – Weakest acid C2H5NH2 + H2O ↔ C2H5NH3 + + OH- 252 352 NHHC OHNHHC Kb NH3 + H2O ↔ NH4 + + OH- 3 4 NH OHNH Kb N2H4 + H2O ↔ N2H5 + + OH- 42 52 HN OHHN Kb Kb – Highest – Strongest base Kb – Lowest – Weakest base Table shows Ka/Kb values for diff substances at 298K. Arrange in order of increasing strength of acidity8 Substance A B C D E pKa/ pKb pKa = 3.4 pKa = 6.7 pKa= 2.1 PKb = 6 pKb= 5 pKa = - lg10Ka pKb = - lg10Kb pKa + pKb = pKw pKa + pKb = 14 Low pKa – High Ka – Strong acid pKa = 14-6 = 8 pKa = 14-5 = 9C > A > B > D > E
- 25. A. 0.3 mol NH3 and 0.3 mol HCI B. 0.3 mol NH3 and 0.15 mol HCI C. 0.3 mol NH3 and 0.6 mol HCI D. 0.3 mol NH3 and 0.15 mol H2SO4 I. 50ml, 0.1M CH3COONa II. 25ml, 0.1M NaOH III. 50ml, 0.1M NaOH IB Questions 9 Adding weak acid and its conjugate base/salt Titrating Buffer preparation CH3COOH / CH3COONa Acidic buffer Basic buffer NH3 / NH4CI How buffer solution are prepared? Weak acid with strong base Weak base with strong acid Weak acid Strong base Strong acid Weak base Buffer can be prepared by adding which of the followingto 50ml, 0.1M CH3COOH10 Which solution will produce a buffer in 1dm3 of water?11 12 Which substance could be added to ethanoic acid to prepare acidic buffer? I. Hydrochloric acid II. Sodium ethanoate III. Sodium hydroxide
- 26. 25ml weak acid, HA titrated with 0.155M NaOH and graph is shown below a) Determine pH at equivalence point b) Explain using eqn, why equivalence point is not at pH = 7 c) Calc conc of weak acid bef addition of any NaOH d) Estimate, using data from graph, Ka of weak acid Sample IB Questionon Acid Base Titration a) pH is = 9 At equivalencepoint Amt acid = Amt base HA M = ? M V = 25.0ml NaOH M = 0.155M V = 22 ml c) HA + NaOH → NaA + H2O Moles of Base = MV = (0.155 x 0.022) = 3.41 x 10-3 Mole ratio ( 1 : 1) • 1 mole base neutralize 1 mole acid • 3.41 x 10-3 base neutralize 3.41 x 10-3 acid Moles Acid = MV = M x 0.025 M x 0.025 = 3.41 x 10-3 M = 0.136M d) pH = 5.3 At half equivalence pt: • Vol base 11ml • Amt acid = amt salt pH = pKa - lg [acid] [salt] pH = pKa = 5.3 pKa = -lg Ka 5.3 = -lg Ka Ka = 5 x 10-6 b) Neutralization - strong base with weak acid HA + NaOH → A- + H2O A- is a strong conjugate base A- + H2O → HA + OH- (basic salt) 3.41 x 10-3 base added Graph below shows variation pH for titration bet 25.0ml H3PO4 with 0.1M NaOH. Write balanced eqn for rxn at pH=4.7, pH = 9.6, pH = 12.5. What vol of NaOH needed to neutralize 25ml H3PO4. 13 11 Click here notes PolyproticAcid – dissociate1 protonstepwise 1st H+ dissociation - H3PO4 ↔ H+ + H2PO4 - 2nd H+ dissociation – H2PO4 - ↔ H+ + HPO4 2- 3rd H+ dissociation – HPO4 2- ↔ H+ + PO4 3- 13 3 108.4 aK Ka get smaller – Weaker acid – Difficult remove H+ from an increasingly negatively charged.Stronger ESF bet H+ and anion pH = 4.7 pH = 9.6 pH = 12.5 3 1 105.7 aK 8 2 102.6 aK
- 27. Click here on titration animationClick here on titration simulation Simulation and Animation on Buffer and Titrations Click here for videos from Khan Academy Click here acidic buffer animationClick here titration animation Click here titration simulation Click here titration animation Click here titration animation
