ARM 2007: Dissection, characterisation and utilisation of disease QTL -- R Nelson
•
1 gostou•4,911 visualizações
Denunciar
Compartilhar
Denunciar
Compartilhar
Baixar para ler offline
Recomendados
ARM 2008: Dissection, characterisation and utilisation of disease QTL -- R Ne...

ARM 2008: Dissection, characterisation and utilisation of disease QTL -- R Ne...CGIAR Generation Challenge Programme
GRM 2013: Improving and deploying markers for biotic stresses in cassava -- C...

GRM 2013: Improving and deploying markers for biotic stresses in cassava -- C...CGIAR Generation Challenge Programme
Recomendados
ARM 2008: Dissection, characterisation and utilisation of disease QTL -- R Ne...

ARM 2008: Dissection, characterisation and utilisation of disease QTL -- R Ne...CGIAR Generation Challenge Programme
GRM 2013: Improving and deploying markers for biotic stresses in cassava -- C...

GRM 2013: Improving and deploying markers for biotic stresses in cassava -- C...CGIAR Generation Challenge Programme
H. ZegeyeGenome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...

Genome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...Borlaug Global Rust Initiative
The introduction of defined Ion AmpliSeq™ panels for detection and characterization of actionable mutations occurring in tumor tissue has the potential to revolutionize translational oncology research. The Ion Ampliseq™ cancer hot spot panel version 2 (CHP v2) by Ion Torrent includes 207 actionable sequences from a single target and mutation targets present in 50 genes and the more comprehensive Ion Oncomine™ cancer panel (OCP) developed by Life Technologies Compendia Bioscience™ contains over 2000 mutations. A hallmark of these Ion Torrent Ampliseq cancer panels is the low amount of input DNA needed which is critical when the clinical specimen material is limited such as with fine needle biopsy or FFPE samples. Typically, 10 ng of DNA obtained from these sources is sufficient to produce informative sequencing data. Often, cancer-causing or promoting mutations are detected at relatively low allele frequencies like 10-20 % compared to the major normal allele. Many researchers wish to verify these findings of low frequency mutations by an orthologous method such as traditional dye-fluorescent Sanger sequencing on a capillary electrophoresis (CE) instrument such as the Applied Biosystems 3500 genetic analyzer. To that end, we have developed a workflow that enables the amplification and traditional Sanger sequencing of individual Ion AmpliSeq targets directly from the AmpliSeq library starting material.
The method requires a retainer of 1 μl (~ 5%) of the original AmpliSeq preamplification material. A dilution of this aliquot is used as template source for individualized PCR/sequencing reactions. We show that a random selection of 48 targets from the CHPv2 panel could be successfully amplified and Sanger-sequenced from an Ion Torrent Ampliseq library originally prepared from 10 ng of FFPE
DNA. Furthermore, we show the successful Sanger-re-sequencing of all individual 24 targets covering the TP53 exons from the same sample processed and pre-amplified with the OncoMine AmpliSeq panel.
Taken together, this method will enable researchers to reflex-test potential mutations of interest from very material-limited specimen using Sanger CE sequencingDirect Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...

Direct Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...Thermo Fisher Scientific
GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...

GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...CGIAR Generation Challenge Programme
GRM2013: Establishing a molecular breeding program based on the aluminum tole...

GRM2013: Establishing a molecular breeding program based on the aluminum tole...CGIAR Generation Challenge Programme
GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...

GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...CGIAR Generation Challenge Programme
Mais conteúdo relacionado
Mais procurados
H. ZegeyeGenome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...

Genome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...Borlaug Global Rust Initiative
The introduction of defined Ion AmpliSeq™ panels for detection and characterization of actionable mutations occurring in tumor tissue has the potential to revolutionize translational oncology research. The Ion Ampliseq™ cancer hot spot panel version 2 (CHP v2) by Ion Torrent includes 207 actionable sequences from a single target and mutation targets present in 50 genes and the more comprehensive Ion Oncomine™ cancer panel (OCP) developed by Life Technologies Compendia Bioscience™ contains over 2000 mutations. A hallmark of these Ion Torrent Ampliseq cancer panels is the low amount of input DNA needed which is critical when the clinical specimen material is limited such as with fine needle biopsy or FFPE samples. Typically, 10 ng of DNA obtained from these sources is sufficient to produce informative sequencing data. Often, cancer-causing or promoting mutations are detected at relatively low allele frequencies like 10-20 % compared to the major normal allele. Many researchers wish to verify these findings of low frequency mutations by an orthologous method such as traditional dye-fluorescent Sanger sequencing on a capillary electrophoresis (CE) instrument such as the Applied Biosystems 3500 genetic analyzer. To that end, we have developed a workflow that enables the amplification and traditional Sanger sequencing of individual Ion AmpliSeq targets directly from the AmpliSeq library starting material.
The method requires a retainer of 1 μl (~ 5%) of the original AmpliSeq preamplification material. A dilution of this aliquot is used as template source for individualized PCR/sequencing reactions. We show that a random selection of 48 targets from the CHPv2 panel could be successfully amplified and Sanger-sequenced from an Ion Torrent Ampliseq library originally prepared from 10 ng of FFPE
DNA. Furthermore, we show the successful Sanger-re-sequencing of all individual 24 targets covering the TP53 exons from the same sample processed and pre-amplified with the OncoMine AmpliSeq panel.
Taken together, this method will enable researchers to reflex-test potential mutations of interest from very material-limited specimen using Sanger CE sequencingDirect Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...

Direct Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...Thermo Fisher Scientific
Mais procurados (19)
Genome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...

Genome-wide Association Mapping of Adult Plant Resistance to Stripe Rust in S...
DEVELOPMENT OF A NEW EXPERT-CURATED FOOT-AND-MOUTH DISEASE VIRUS NUCLEOTIDE S...

DEVELOPMENT OF A NEW EXPERT-CURATED FOOT-AND-MOUTH DISEASE VIRUS NUCLEOTIDE S...
12 apple characteristics of prebreeding material kellerhals markus

12 apple characteristics of prebreeding material kellerhals markus
READY-TO-USE SOLID PHASE BLOCKING ELISA KITS FOR DETECTION OF SPECIFIC ANTIBO...

READY-TO-USE SOLID PHASE BLOCKING ELISA KITS FOR DETECTION OF SPECIFIC ANTIBO...
CNV and aneuploidy detection by Ion semiconductor sequencing

CNV and aneuploidy detection by Ion semiconductor sequencing
Rapid Detection of Aneuploidy from Multiplexed Single Cell Samples

Rapid Detection of Aneuploidy from Multiplexed Single Cell Samples
Validation of shoot fly resistance introgression lines using SNP markers in S...

Validation of shoot fly resistance introgression lines using SNP markers in S...
Cytoscan_Copy_Number_Confirmation_with_SYBR_Green_qPCR_white_paper

Cytoscan_Copy_Number_Confirmation_with_SYBR_Green_qPCR_white_paper
Deep Sequencing Identifies Novel Circulating and Hepatic ncRNA Profiles in NA...

Deep Sequencing Identifies Novel Circulating and Hepatic ncRNA Profiles in NA...
Results from a Interlaboratory exercise to evaluate NSP kits (C. Browning)

Results from a Interlaboratory exercise to evaluate NSP kits (C. Browning)
Blue tongue virus in Kenya: Insights from Isiolo and progress on molecular ty...

Blue tongue virus in Kenya: Insights from Isiolo and progress on molecular ty...
Direct Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...

Direct Sanger CE Sequencing of Individual Ampliseq Cancer Panel Targets from ...
iDiffIR: Identifying differential intron retention from RNA-seq

iDiffIR: Identifying differential intron retention from RNA-seq
Destaque
GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...

GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...CGIAR Generation Challenge Programme
GRM2013: Establishing a molecular breeding program based on the aluminum tole...

GRM2013: Establishing a molecular breeding program based on the aluminum tole...CGIAR Generation Challenge Programme
GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...

GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...CGIAR Generation Challenge Programme
GRM 2013: Sorghum product catalogue and project status -- Projects ongoing, c...

GRM 2013: Sorghum product catalogue and project status -- Projects ongoing, c...CGIAR Generation Challenge Programme
GRM 2013: Marker-assisted breeding for improving phosphorus-use efficiency an...

GRM 2013: Marker-assisted breeding for improving phosphorus-use efficiency an...CGIAR Generation Challenge Programme
GRM 2013: Developing drought-adapted sorghum germplasm for Africa and Austral...

GRM 2013: Developing drought-adapted sorghum germplasm for Africa and Austral...CGIAR Generation Challenge Programme
GRM 2011: Development and evaluation of drought-adapted sorghum germplasm for...

GRM 2011: Development and evaluation of drought-adapted sorghum germplasm for...CGIAR Generation Challenge Programme
These slides are a redacted (by the presenter) version of what was presented at the meetingGRM 2013: Validation of ZmMATEs as Genes Underlying Major Al Tolerance QTLs i...

GRM 2013: Validation of ZmMATEs as Genes Underlying Major Al Tolerance QTLs i...CGIAR Generation Challenge Programme
GRM 2013: Cloning, characterization and validation of AltSB/Al tolerance in r...

GRM 2013: Cloning, characterization and validation of AltSB/Al tolerance in r...CGIAR Generation Challenge Programme
GRM 2013: Status of maize projects in the Comparative Genomics Research Init...

GRM 2013: Status of maize projects in the Comparative Genomics Research Init...CGIAR Generation Challenge Programme
GRM 2013: Cloning, characterization and validation of PUP1/P efficiency in ma...

GRM 2013: Cloning, characterization and validation of PUP1/P efficiency in ma...CGIAR Generation Challenge Programme
GRM 2013: Asian Maize Drought Tolerance (AMDROUT) Project -- BS Vivek

GRM 2013: Asian Maize Drought Tolerance (AMDROUT) Project -- BS VivekCGIAR Generation Challenge Programme
GRM 2011: KEYNOTE ADDRESS-2: Integrated Breeding -- Impact and challenges for...

GRM 2011: KEYNOTE ADDRESS-2: Integrated Breeding -- Impact and challenges for...CGIAR Generation Challenge Programme
These slides are a redacted (by the presenter) version of what was presented at the meetingGRM 2013: Improving phosphorus efficiency in sorghum by the identification an...

GRM 2013: Improving phosphorus efficiency in sorghum by the identification an...CGIAR Generation Challenge Programme
GRM 2013: Improve cowpea productivity for marginal environments in sub-Sahar...

GRM 2013: Improve cowpea productivity for marginal environments in sub-Sahar...CGIAR Generation Challenge Programme
PAG XXII 2014 – Genomic resources applied to marker-assisted breeding in cowp...

PAG XXII 2014 – Genomic resources applied to marker-assisted breeding in cowp...CGIAR Generation Challenge Programme
Ravi Singh, CIMMYTPros and cons of utilizing major, race-specific resistance genes versus parti...

Pros and cons of utilizing major, race-specific resistance genes versus parti...Borlaug Global Rust Initiative
Destaque (20)
GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...

GRM 2013: Enhancing sorghum grain yield and quality for the Sudano-Sahelian z...
GRM2013: Establishing a molecular breeding program based on the aluminum tole...

GRM2013: Establishing a molecular breeding program based on the aluminum tole...
GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...

GRM 2013: Improving sorghum productivity in semi-arid environments of Mali th...
GRM 2013: Sorghum product catalogue and project status -- Projects ongoing, c...

GRM 2013: Sorghum product catalogue and project status -- Projects ongoing, c...
GRM 2013: Marker-assisted breeding for improving phosphorus-use efficiency an...

GRM 2013: Marker-assisted breeding for improving phosphorus-use efficiency an...
GRM 2013: Developing drought-adapted sorghum germplasm for Africa and Austral...

GRM 2013: Developing drought-adapted sorghum germplasm for Africa and Austral...
GRM 2011: Development and evaluation of drought-adapted sorghum germplasm for...

GRM 2011: Development and evaluation of drought-adapted sorghum germplasm for...
GRM 2011: Asian Maize Drought Tolerance (AMDROUT) Project

GRM 2011: Asian Maize Drought Tolerance (AMDROUT) Project
GRM 2013: Validation of ZmMATEs as Genes Underlying Major Al Tolerance QTLs i...

GRM 2013: Validation of ZmMATEs as Genes Underlying Major Al Tolerance QTLs i...
GRM 2013: Cloning, characterization and validation of AltSB/Al tolerance in r...

GRM 2013: Cloning, characterization and validation of AltSB/Al tolerance in r...
GRM 2013: Status of maize projects in the Comparative Genomics Research Init...

GRM 2013: Status of maize projects in the Comparative Genomics Research Init...
GRM 2013: Cloning, characterization and validation of PUP1/P efficiency in ma...

GRM 2013: Cloning, characterization and validation of PUP1/P efficiency in ma...
GRM 2013: Asian Maize Drought Tolerance (AMDROUT) Project -- BS Vivek

GRM 2013: Asian Maize Drought Tolerance (AMDROUT) Project -- BS Vivek
GRM 2011: KEYNOTE ADDRESS-2: Integrated Breeding -- Impact and challenges for...

GRM 2011: KEYNOTE ADDRESS-2: Integrated Breeding -- Impact and challenges for...
TLI 2012: Data management for chickpea breeding - Kenya

TLI 2012: Data management for chickpea breeding - Kenya
GRM 2013: Improving phosphorus efficiency in sorghum by the identification an...

GRM 2013: Improving phosphorus efficiency in sorghum by the identification an...
GRM 2013: Improve cowpea productivity for marginal environments in sub-Sahar...

GRM 2013: Improve cowpea productivity for marginal environments in sub-Sahar...
PAG XXII 2014 – Genomic resources applied to marker-assisted breeding in cowp...

PAG XXII 2014 – Genomic resources applied to marker-assisted breeding in cowp...
Pros and cons of utilizing major, race-specific resistance genes versus parti...

Pros and cons of utilizing major, race-specific resistance genes versus parti...
Semelhante a ARM 2007: Dissection, characterisation and utilisation of disease QTL -- R Nelson
MCO 2011 - Slide 30 - K. Öberg - Spotlight session - Neuroendocrine tumours

MCO 2011 - Slide 30 - K. Öberg - Spotlight session - Neuroendocrine tumoursEuropean School of Oncology
ECCLU 2011 - A. Bex - Kidney cancer - Adjuvant and neo-adjuvant treatment

ECCLU 2011 - A. Bex - Kidney cancer - Adjuvant and neo-adjuvant treatmentEuropean School of Oncology
Semelhante a ARM 2007: Dissection, characterisation and utilisation of disease QTL -- R Nelson (20)
09 Tratamiento combinado de quimio y radioterapia en Cáncer de Pulmón

09 Tratamiento combinado de quimio y radioterapia en Cáncer de Pulmón
Use of Affymetrix Arrays (GeneChip® Human Transcriptome 2.0 Array and Cytosca...

Use of Affymetrix Arrays (GeneChip® Human Transcriptome 2.0 Array and Cytosca...
SCS macrophages suppress melanoma by restricting tumor-derived vesicle–B cell...

SCS macrophages suppress melanoma by restricting tumor-derived vesicle–B cell...
Eligibility for ACOSOG Z0011 Trial and Results on a Cohort of 3546 Breast Can...

Eligibility for ACOSOG Z0011 Trial and Results on a Cohort of 3546 Breast Can...
From syngenic to_humanised_models_mirjolet_oncodesign

From syngenic to_humanised_models_mirjolet_oncodesign
MCO 2011 - Slide 30 - K. Öberg - Spotlight session - Neuroendocrine tumours

MCO 2011 - Slide 30 - K. Öberg - Spotlight session - Neuroendocrine tumours
ECCLU 2011 - A. Bex - Kidney cancer - Adjuvant and neo-adjuvant treatment

ECCLU 2011 - A. Bex - Kidney cancer - Adjuvant and neo-adjuvant treatment
Jiankang Wang. Principle of QTL mapping and inclusive composite interval mapp...

Jiankang Wang. Principle of QTL mapping and inclusive composite interval mapp...
Eligibility for ACOSOG Z0011 Trial and Results on a Cohort of 3546 Breast Can...

Eligibility for ACOSOG Z0011 Trial and Results on a Cohort of 3546 Breast Can...
Mais de CGIAR Generation Challenge Programme
Presented by the GCP Director at the
Tropical Soybean for Development Workshop Meeting
28th October 2014
Washington DC, USA
Capacity Building: Gain or Drain? J-M Ribaut, F Okono and NN Diop

Capacity Building: Gain or Drain? J-M Ribaut, F Okono and NN DiopCGIAR Generation Challenge Programme
Presentation made by the GCP Director during the CGIAR Fund Council (FC) visit to CIMMYT (GCP's host), on the sidelines of the FC meeting in Mexico in May 2014.The Generation Challenge Programme: Lessons learnt relevant to CRPs, and the ...

The Generation Challenge Programme: Lessons learnt relevant to CRPs, and the ...CGIAR Generation Challenge Programme
Slides presented at University of Eldoret, Kenya, including faculty staff and students from Moi UniversityIntegrated Breeding Platform (IBP): A user-friendly platform to implement the...

Integrated Breeding Platform (IBP): A user-friendly platform to implement the...CGIAR Generation Challenge Programme
Slides presented at Egerton University, Njoro, Kenya, in March 2014Integrated Breeding Platform (IBP): A user-friendly platform to implement the...

Integrated Breeding Platform (IBP): A user-friendly platform to implement the...CGIAR Generation Challenge Programme
These slides were presented by Clare MukankusiTLM III: : Improve common bean productivity for marginal environments in su...

TLM III: : Improve common bean productivity for marginal environments in su...CGIAR Generation Challenge Programme
TLM III: Improve groundnut productivity for marginal environments from sub-Sa...

TLM III: Improve groundnut productivity for marginal environments from sub-Sa...CGIAR Generation Challenge Programme
TLM III: Improve cowpea productivity for marginal environments in sub-Sahara...

TLM III: Improve cowpea productivity for marginal environments in sub-Sahara...CGIAR Generation Challenge Programme
TLIII: Overview of TLII achievements, lessons and challenges for Phase III – ...

TLIII: Overview of TLII achievements, lessons and challenges for Phase III – ...CGIAR Generation Challenge Programme
TLIII: Tropical Legumes I – Improving Tropical Legume Productivity for Margin...

TLIII: Tropical Legumes I – Improving Tropical Legume Productivity for Margin...CGIAR Generation Challenge Programme
Presentation by the GCP Director at an international workshop on genomics and integrated breeding, February 2014. More on the workshop: http://bit.ly/MwpliD You can also view the presentation on video here: http://bit.ly/1mVmVdS Adoption of modern breeding tools in developing countries: challenges and opp...

Adoption of modern breeding tools in developing countries: challenges and opp...CGIAR Generation Challenge Programme
PAG XXII 2014 – The Breeding Management System (BMS) of the Integrated Breedi...

PAG XXII 2014 – The Breeding Management System (BMS) of the Integrated Breedi...CGIAR Generation Challenge Programme
PAG XXII 2014 – The Crop Ontology: A resource for enabling access to breeders...

PAG XXII 2014 – The Crop Ontology: A resource for enabling access to breeders...CGIAR Generation Challenge Programme
A quick introduction to the CGIAR Generation Challenge Programme (GCP) -- its history, network, research organisation, outputs and challenges. GCP is a virtual network of partnerships working on modern crop breeding for food security2011: Introduction to the CGIAR Generation Challenge Programme (GCP)

2011: Introduction to the CGIAR Generation Challenge Programme (GCP)CGIAR Generation Challenge Programme
Presentation by GCP Director to plant industry business executives attending Limagrain Annual Meeting, Faro, Portugal, January 23, 2013
Working with diversity in international partnerships -- The GCP experience --...

Working with diversity in international partnerships -- The GCP experience --...CGIAR Generation Challenge Programme
GRM 2013: Improving rice productivity in lowland ecosystems of Burkina Faso, ...

GRM 2013: Improving rice productivity in lowland ecosystems of Burkina Faso, ...CGIAR Generation Challenge Programme
GRM 2013: Wheat product catalogue and project status -- Projects ongoing, com...

GRM 2013: Wheat product catalogue and project status -- Projects ongoing, com...CGIAR Generation Challenge Programme
GRM 2013: Rice product catalogue and project status -- Projects ongoing, comp...

GRM 2013: Rice product catalogue and project status -- Projects ongoing, comp...CGIAR Generation Challenge Programme
GRM 2013: Groundnut product catalogue and project status -- Projects ongoing,...

GRM 2013: Groundnut product catalogue and project status -- Projects ongoing,...CGIAR Generation Challenge Programme
GRM 2013: Cowpea product catalogue and project status -- Projects ongoing, co...

GRM 2013: Cowpea product catalogue and project status -- Projects ongoing, co...CGIAR Generation Challenge Programme
Mais de CGIAR Generation Challenge Programme (20)
Capacity Building: Gain or Drain? J-M Ribaut, F Okono and NN Diop

Capacity Building: Gain or Drain? J-M Ribaut, F Okono and NN Diop
The Generation Challenge Programme: Lessons learnt relevant to CRPs, and the ...

The Generation Challenge Programme: Lessons learnt relevant to CRPs, and the ...
Lessons learnt from the GCP experience – J-M Ribaut

Lessons learnt from the GCP experience – J-M Ribaut
Integrated Breeding Platform (IBP): A user-friendly platform to implement the...

Integrated Breeding Platform (IBP): A user-friendly platform to implement the...
Integrated Breeding Platform (IBP): A user-friendly platform to implement the...

Integrated Breeding Platform (IBP): A user-friendly platform to implement the...
TLM III: : Improve common bean productivity for marginal environments in su...

TLM III: : Improve common bean productivity for marginal environments in su...
TLM III: Improve groundnut productivity for marginal environments from sub-Sa...

TLM III: Improve groundnut productivity for marginal environments from sub-Sa...
TLM III: Improve cowpea productivity for marginal environments in sub-Sahara...

TLM III: Improve cowpea productivity for marginal environments in sub-Sahara...
TLIII: Overview of TLII achievements, lessons and challenges for Phase III – ...

TLIII: Overview of TLII achievements, lessons and challenges for Phase III – ...
TLIII: Tropical Legumes I – Improving Tropical Legume Productivity for Margin...

TLIII: Tropical Legumes I – Improving Tropical Legume Productivity for Margin...
Adoption of modern breeding tools in developing countries: challenges and opp...

Adoption of modern breeding tools in developing countries: challenges and opp...
PAG XXII 2014 – The Breeding Management System (BMS) of the Integrated Breedi...

PAG XXII 2014 – The Breeding Management System (BMS) of the Integrated Breedi...
PAG XXII 2014 – The Crop Ontology: A resource for enabling access to breeders...

PAG XXII 2014 – The Crop Ontology: A resource for enabling access to breeders...
2011: Introduction to the CGIAR Generation Challenge Programme (GCP)

2011: Introduction to the CGIAR Generation Challenge Programme (GCP)
Working with diversity in international partnerships -- The GCP experience --...

Working with diversity in international partnerships -- The GCP experience --...
GRM 2013: Improving rice productivity in lowland ecosystems of Burkina Faso, ...

GRM 2013: Improving rice productivity in lowland ecosystems of Burkina Faso, ...
GRM 2013: Wheat product catalogue and project status -- Projects ongoing, com...

GRM 2013: Wheat product catalogue and project status -- Projects ongoing, com...
GRM 2013: Rice product catalogue and project status -- Projects ongoing, comp...

GRM 2013: Rice product catalogue and project status -- Projects ongoing, comp...
GRM 2013: Groundnut product catalogue and project status -- Projects ongoing,...

GRM 2013: Groundnut product catalogue and project status -- Projects ongoing,...
GRM 2013: Cowpea product catalogue and project status -- Projects ongoing, co...

GRM 2013: Cowpea product catalogue and project status -- Projects ongoing, co...
Último
Último (20)
TEST BANK For Radiologic Science for Technologists, 12th Edition by Stewart C...

TEST BANK For Radiologic Science for Technologists, 12th Edition by Stewart C...
Labelling Requirements and Label Claims for Dietary Supplements and Recommend...

Labelling Requirements and Label Claims for Dietary Supplements and Recommend...
PossibleEoarcheanRecordsoftheGeomagneticFieldPreservedintheIsuaSupracrustalBe...

PossibleEoarcheanRecordsoftheGeomagneticFieldPreservedintheIsuaSupracrustalBe...
GUIDELINES ON SIMILAR BIOLOGICS Regulatory Requirements for Marketing Authori...

GUIDELINES ON SIMILAR BIOLOGICS Regulatory Requirements for Marketing Authori...
Formation of low mass protostars and their circumstellar disks

Formation of low mass protostars and their circumstellar disks
Creating and Analyzing Definitive Screening Designs

Creating and Analyzing Definitive Screening Designs
Asymmetry in the atmosphere of the ultra-hot Jupiter WASP-76 b

Asymmetry in the atmosphere of the ultra-hot Jupiter WASP-76 b
SAMASTIPUR CALL GIRL 7857803690 LOW PRICE ESCORT SERVICE

SAMASTIPUR CALL GIRL 7857803690 LOW PRICE ESCORT SERVICE
Pulmonary drug delivery system M.pharm -2nd sem P'ceutics

Pulmonary drug delivery system M.pharm -2nd sem P'ceutics
Vip profile Call Girls In Lonavala 9748763073 For Genuine Sex Service At Just...

Vip profile Call Girls In Lonavala 9748763073 For Genuine Sex Service At Just...
Hire 💕 9907093804 Hooghly Call Girls Service Call Girls Agency

Hire 💕 9907093804 Hooghly Call Girls Service Call Girls Agency
Disentangling the origin of chemical differences using GHOST

Disentangling the origin of chemical differences using GHOST
Kochi ❤CALL GIRL 84099*07087 ❤CALL GIRLS IN Kochi ESCORT SERVICE❤CALL GIRL

Kochi ❤CALL GIRL 84099*07087 ❤CALL GIRLS IN Kochi ESCORT SERVICE❤CALL GIRL
Hubble Asteroid Hunter III. Physical properties of newly found asteroids

Hubble Asteroid Hunter III. Physical properties of newly found asteroids
VIRUSES structure and classification ppt by Dr.Prince C P

VIRUSES structure and classification ppt by Dr.Prince C P
Stunning ➥8448380779▻ Call Girls In Panchshil Enclave Delhi NCR

Stunning ➥8448380779▻ Call Girls In Panchshil Enclave Delhi NCR
ARM 2007: Dissection, characterisation and utilisation of disease QTL -- R Nelson
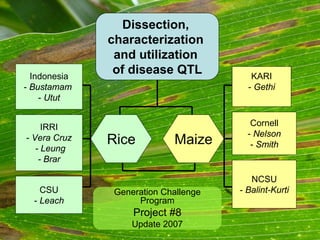
- 1. Indonesia-Bustamam -UtutIRRI-Vera Cruz-Leung-BrarCSU-LeachKARI-GethiCornell-Nelson-SmithNCSU-Balint-KurtiRiceMaizeGeneration Challenge ProgramProject #8Update 2007Dissection, characterization and utilization of disease QTL
- 2. Synthesis maps(Complementary) mechanismsInsight on MDRUnderstanding the traitGermplasm panel: resistance sourcesStress resistant varieties(Actionable) insightsre: breeding populationsImproving the traitdQTLdiscoverydQTLgenesDissecting the traitdQTLvs. expressiondQTLNILsPublishedNILs; HistopathologyDiversity panel; RS populationsMapping populationsDisease panelAssociation panelDiallelSelection from diallelSelection from AB- QTL populations(poster 3.25) Linkage mapping Selection mappingAB-QTL mappingMutant analysisMultiple strategies(poster 2.9) Deletion analysisQTL dissectionAssocationmappingNAMArray studies
- 3. GR6458GR6489GR6603GR6686GR7534GR6728GR6989Chip-analysis of loss-of-resistance mutantsJ. Leach, G. Diaz, and M. Bruce, CSUspl1LOC_Os01g27150LOC_Os03g56800LOC_Os04g54240 IR64IR64IR64GR6686GR6603GR6603GR7534GR6728GR6686 LOC_Os01g64720 GR6603GR6686GR6728GR7534IR64 LOC_Os01g67720 GR6603GR6686GR6728GR6989IR64GR7534
- 4. Allelic series of deletion mutants Gene model position on chromosome 12 Ratio of significant probes in probe set Mutant lineAll mutants show spl1 phenotype. Two had been confirmed as allelic by complementation tests
- 5. OXLP clusterOxalate oxidase(germin-like protein) R2 = 0.7039P = <0.0010.00.51.01.52.002468Rice disease rating Relative expression T0/WT Rice blast disease ratingMore More silencingsilencingCarilloand Vera Cruz, IRRIRNAi Manosalvaand Leach, CSU Increased silencing of Chr. 8 OsGLPgenes in T0 rice correlates with increased disease
- 6. u1228m0371Chromosome 4p072u1008b1434nc135u1669u1509b1126u2281p096b1265u2054u1142u1451u1896u2027u2038b1189u1808u2041m0321u2285u1313u1999u1101b589b18900.050100150200250300350400450500550600650700Chromosome 1Chromosome 2p96100u2246u2363b1017b1338m0111u1756u1261u2248p109642b1018m0401b1831b1036b1138u1080u2372b1329u1042b1662b1940b1520u1252p101049u1696AY1112360.050100150200250300350400450500550600650700Chromosome 3u2118u2256p104127u1886b1325b1523u1057u2258u2369p099u2000u2158b2047u2261u2264u1773b1456b1904u1920u2376u1266b1449b1047u2266u2269 u2270u2271dupssr17b1605u1825m0251b1108u1273u2276dupssr33b1182b1496u1062u1594u11360.050100150200250300350400450500550600650700750800Chromosome 5u1308u1496u1781b105b1046u1447u1355b1287b1208m0081u1853u1722p101u2216p048b2305b118AY105910u23070.050100150200250300350400450500550600650Chromosome 10p041u1293u1291u1152u2034u1337p059u1863u1866p050u1995b1526u1077u1115u1506b1028u1084b1360b1185501001502002503003504004500.0b1065Chromosome 8u1414b1194u1913b1834b1863u1457b1176b666b1782b1031dupssr14b1056b11310.050100150200250300350400450500550600Chromosome 6b1043b426b1371u1083u1572AY111964m0241b1732u1859p299852b1740b1521b1538nc009u23240.050100150200250300350400450500Chromosome 7b1367b2132u1159b1094u1983b1022b1808b1305b1070b1805b1666b2259b1407p0510.050100150200250300350400450500550600Chromosome 98000.050100150200250300350400450500550-39.0700750600650u1279b1724u1867u2393b1810b1583u2335u1596u1430b244p061u1107u2398u1120u1078u1231u2095u2343u2371b1191u1366b619b128 u1982u1277NC250 Introgression likely derived from B37NC250 Introgression likely derived from Nigerian Composite ARbNC250 Introgression in both NC292 and NC330NC250 Introgression in NC292NC250 Introgression in NC330Unknown no SSR dataUnknown bands present in NC292 and NC330 not present in B73 or NC250Unknown bands present NC330 not present in B73 or NC250, NC292 has B73 bandMajor SLB QTL positions B73/NC250A F2:3populationumc1704umc1532umc2011bnlg1917u1636dupssr19b2228u1177b1490.050a31b1429u1160b1484b1866b652u2227b2086u1297u1988u1254u1035u1122u1661u1358b1556b1025u1128p037u1848m0041u2047u1298u1431b1671b1347u1500p265454u1553u1129u1118b2123u1725u2244100150200250300350400450500550600650700750800850900950100010501100u2053u1576b594IDP1471IDP4016500BBCCGGFFEEDDIIJJHHAAIntrogression CIntrogression FIntrogression GIntrogression JB73NC292Introgression BIntrogression DZwonitzer and Balint-Kurti
- 7. Increase in rate of lesion development0.070.0750.080.0850.090.095B73HetSEL* Incubation Period -1Antagonistic reactions for NLB and rust resistance0102030405060B73HetSEL* DLADecrease in rust severity R SRSSelected alleleNLBp < 0.001Rustp = 0.01 J.A. Poland and R.J. Nelson, Cornell
- 8. Chung and NelsonIncidence of E. turcicum growing into thexylem on different maize genotypes(7 days after inoculation) 020406080100CML52Tx303B73TBBC3_42Maize genotype % of infection sites hyphae in the xylem
- 9. 0102030405060708090100110 IFKi3M162W20602511CML69201CML322B7312817378171219127119Ky21484281M37W26CML322-Inoc192194185137Tx3037990615466501661968CML322-MockOh7B Line Infection Frequency (%) Maize + drought = aflatoxicosis Miderosand Nelson, CornellResistance to infection by Aspergillusflavus
- 10. Chr 2Chr 3Chr 5Chr 7Panicle weightLeaf blastLesion number Diseased leaf area Harvest indexHeightGrain fillingMapping of QTLsfor resistance to blast and drought tolerance in rice Vandana/ Moroberekanderivatives; IRRI & IndiaDatasets and lines: Advanced rice genotypes with drought and blast resistanceMoroberekan/ Vandana(India-IRRI) Oryzicallanos 5 / Way Rarem(Indonesia / IRRI)
- 11. Genetics + breeding Parent 1&2 CML 312 CML 384 TZMi 102 TZMi 711 TZMi 712 CML 204 CML 373 CML 312 Self CML 384 Self TZMi 102 Self TZMi 711 Self TZMi 712 Self CML 204 Self CML 373 SelfModerate x ModerateCML 312 x TZMi 712 •Diallelstudy of GLS resistance •F2:3and BC4F4lines pending •Promising hybrids identified
- 12. Gene(s) QDR-QTLQTL Dissection NILsNested Association MappingAssociationmapping RIL populations Selection mapping Targeted HIF extraction Introgression librariesDeletion mutantsVarietiesAffymetrixdata on deletion mutants5 SLB datasets4 GLS datasets 3 NLB datasets2 SLB datasets2 NLB datasets2 smut datasets(collabwith Buckler / Holland) Materials, marker data and phenodata
