Computational Modeling of Biophysical Processes in a Cell
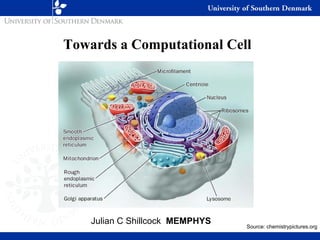
- 1. Towards a Computational Cell Julian C Shillcock MEMPHYS Source: chemistrypictures.org
- 2. Structure of talk What are the organizational and dynamic properties of membranes at a molecular level? How are molecules trafficked among the organelles of a cell? To answer these questions, we could reconstitute model systems in vitro or we can build mathematical and computational models. • Lipids + Water + Proteins + Self-Assembly = Life • Q. Whither DPD Simulations? • Case Study 1: Vesicles and Fusion • Case Study 2: Nanoparticles and Endocytosis • Ans. Simulations that do what you tell them • Conclusions MEMPHYS 2
- 3. Evolution of (Bio-) simulations Past Assembly – random mixture or a few structures (essentially a passive view of the system; we can prepare it but we cannot subsequently interact with it) Present Response – equilibrium properties & perturbations Future Control – we want to interact with a system as it evolves, keep only molecular details necessary to create structure on the scales of interest, observe self-organization and emergent phenomena (The Middle Way Laughlin et al., PNAS 97:32-37, 2000) MEMPHYS 3
- 4. Why not do Molecular Dynamics? • Atomistic Molecular Dynamics is accurate at atomic length-scale (but less useful for macroscopic properties such as shape fluctuations, rigidity,…) • Complex force fields capture motion at short time- scale (bond vibrations, but probably irrelevant for large supramolecular aggregates) Atoms are not the whole story; there are organizing principles above the atomic length scale Fusion event (0.32 µsec. ) with DPD ~200 cpu-hours Fusion event using all-atom MD ~500 cpu-years 4
- 5. Complex Fluids “Simple” fluids are isotropic “Complex” fluids have structure arising out of the “shape” of their constituent molecules, e.g., liquid crystals or lipids with different headgroups or tail lengths Multiple length and time scales, e.g., lipid bilayers have a membrane thickness of ~ 4 nm but form vesicles/cells with diameters from 50 nm to 10 µm; lipids diffuse in ~100 ns but cells Source: chemistrypictures.org divide in ~minutes. Plasma membrane and transport vesicles are composed of hundreds of types of lipid and protein molecules MEMPHYS 5
- 6. Lipids Lipid molecules are amphiphiles and surfactants (surface-active agents) - Water-loving headgroup (1) - Water-hating hydrocarbon tails (2) When placed in water, lipids aggregate into distinct forms: micelle, vesicle, etc. Aggregation is driven by the hydrophobic effect: tendency of water to sequester oily materials so as to maintain its H-bonding network. Properties of the aggregates depend on physical characteristics of lipid molecules, e.g., their “shape”, headgroup size, tail length, as well as their chemical structure. Source: Wikipedia MEMPHYS 6
- 7. Amphiphiles How do we represent amphiphiles in a simulation? Two aspects: - Chemical nature: polar headgroups bound to oily tail(s) - Molecular shape: large or small head/straight or kinked tails Molecular structure leads to a preferred shape in amphiphilic aggregates Inverted Cone Cylinder Cone Source: chemistrypictures.org MEMPHYS 7
- 8. Headgroup Size Amphiphile architecture HC6 4 H2C6 3 H3C6 2 σr 02/kBT 1 0 0.6 0.7 0.8 0.9 -1 -2 -3 Apr/Nr02 -4 Amphiphile architecture modulates planar bilayer response to tension MEMPHYS 8
- 9. Bilayer Self-assembly in Water 324 lipid molecules in (invisible) water Hydrophilic headgroup Hydrocarbon tails Simulation Notes Water is present in all movies, but invisible to reveal dynamics of processes. Periodic Boundary Conditions are used, which means that a molecule leaving one face of the simulation box re-enters at the opposite face. MEMPHYS 9
- 10. Polymer Micelle Self-assembly PEO-PEE diblocks in water: 600 PEO30PEE40 polymers 68 PEO30PEE08 polymers (water invisible) Box = 35 x 35 x 105 nm3 Time = 8 µsec Simulation took 66 cpu-days Self-assembly is a generic property of amphiphiles: different types of aggregate are formed depending on: molecular size, ratio of philic to phobic segments, etc.
- 11. Nanoparticle Self-assembly 216 discoidal nanoparticles (blue) in a Topo /water mixture (7 mM) 4764 Trioctylphosphine (Topo, red/orange) molecules (157 mM)) (Water invisible) Box = (36 nm)3 Simulation took 7 cpu-days Nanoparticle surface is functionalised to bind to Topo headgroup; tails are hydrophobic
- 12. Vesicles Problem of scale: Vesicle area ~ D2 Vesicle volume ~ D3 D = vesicle diameter ~50-500 nm T = membrane thickness ~ 5 nm For realistic vesicle/cell sizes, we need D/T ~ 10-2000. This requires ~800,000 beads for 50 nm vesicle simulation (D/T = 10). A 10 µm cell simulation needs > 1,000,000,000 beads. Current limit is ~ 3,000,000. 9000 lipids in whole membrane; 546 in patch Identical molecular architecture, but different lipid types repel creating a line tension around the patch MEMPHYS 12
- 13. DPD “State of the Art” Applications Polymeric fluids on ~50 nm length scale / microseconds Vesicle fusion ~ 100 nm / microseconds Nanoparticle-membrane interactions: tens of nanoparticles and 50 nm membrane patches Requirements ½ kB per bead of RAM required 1010 bead-steps per cpu-day System size limit is ~3 million particles on single processor: Single fusion event requires ~ 1 cpu-week MEMPHYS 13
- 14. Future Requirements Applications Rational design of drug delivery vehicles Toxicity testing of < 1 µm particles for diagnostics Cell signalling network: receptors, membrane, cytoskeleton, proteins Scales We need: 1 nm – 10 µm, ns – ms We need at least 3 billion particles for a (1 µm)3 run (1 µm)3 for 10 µs requires 274 cpu-years on a single processor: on 1000 nodes with a factor of 1000 speedup, this becomes 0.1 cpu-day and will create ~500 GB per run Hardware/Software Multi-scale model of a cell signalling 1000 commodity, Intel Woodcrest processors; network: fast interconnects; database to hold 100 TB data; XML-based simulation markup language to tag, archive R1 Dissipative Particle Dynamics and re-use simulation results; R2 Brownian Dynamics automated model phase space search R3 Differential equations MEMPHYS 14
- 15. DPD algorithm: Basics Particle based: N particles in a box, specify ri(t) and pi(t), i = 1…N. Mesoscopic: Each particle represents a small volume of fluid with mass, position and momentum Newton’s Laws: Particles interact with surrounding particles; integrate Newton’s equations of motion Three types of force exist between all particles: •Conservative FCij(rij) = aij(1 – |rij|/r0)rij / |rij| FDij(rij) = – γij(1 – |rij|/r0)2(rij.vij) rij / |rij|2 •Dissipative •Random FRij(rij) = (1 – |rij|/r0)ζijrij / |rij| forces are soft, short-ranged (vanish beyond r0), central, pairwise-additive, and conserve momentum locally. MEMPHYS 15
- 16. DPD algorithm: Forces •Conservative FCij(rij) = aij(1 – rij/r0)rij / rij •Dissipative FDij(rij) = – γij(1 – rij/r0)2(rij.vij) rij / rij2 •Random FRij(rij) = (1 – rij/r0)ζijrij / rij Conservative force gives particles an identity, e.g. hydrophobic Dissipative force destroys relative momentum between pairs of interacting particles Random force creates relative momentum between pairs of interacting particles: <ζij (t)> = 0, < ζij (t1) ζij(t2)> = σij2δ(t1-t2), but note that ζij (t) = ζji (t). MEMPHYS 16
- 17. DPD algorithm: Bonds DPD Polymers are constructed by tying particles together with a quadratic potential (Hookean spring): the force law is F(rii+1) = -k2(| rii+1 | - ri0) rii+1 /| rii+1 | with i,i+1 representing adjacent particles in polymer. Note that k2,r0 may depend on the particle types. Hydrocarbon chain stiffness may be included via a bending potential i j V(ijk) = k3(1 - cosφijk) With ijk representing adjacent triples of beads. k Again, k3 may depend on particle types. MEMPHYS 17
- 18. Vesicle Fusion in Cells (Scales et al. Nature 407:144- 146 (2000)). Synaptic vesicles are guided to the pre-synaptic membrane by “motors” moving along filaments; they are then held by SNARE proteins in close proximity to the target membrane. SNAREs hold the vesicle close to the membrane and promote fusion (Knecht & Grubmueller, Biophys. J. 84:1527- 1547(2003)). MEMPHYS 18
- 19. Fusion Protocol: Tension Create bilayer and vesicle under tension 30 nm 50 nm position them close together and let evolve 19
- 20. Fusion Run Vesicle has 5887 lipids; membrane has 5315 in a box (50 nm)3 for 640 ns 20
- 21. Fusion Run Vesicle has 6000 lipids; membrane has 3600 in a box (42 nm)3 for 3.2 µs Lipid headgroup/tail interactions modified to produce a “cone-like” lipid. 21
- 22. Morphology Diagram Bilayer and vesicle lipids: H3(T4)2 Relaxed Nves = 6542 Relaxed Nbil = 8228 43 successful fusion events out of 92 attempts MEMPHYS 22
- 23. Tense Fusion Summary Fusion occurs (within 2 microsec) near the stability limits of the aggregates for this parameter set Our new parameter set shows that flip-flop of lipids from vesicle to planar membrane is one of two time-scales: there are two barriers to fusion: Transfer of vesicle lipids to planar membrane Rearrangement of disordered contact zone into single membrane which subsequently ruptures Shillcock and Lipowsky, Nature Mat. 4:225 (2005) Grafmueller, Shillcock and Lipowsky, PRL 98:218101 (2007) MEMPHYS 23
- 24. Fusion Proteins in vivo SNARE proteins present in both membranes pull them together and drive the formation of the fusion pore But… what do they actually do? Force, torque, displacement…? Do they pull the pore open or prevent it closing? MEMPHYS 24
- 25. Fusion Proteins in silico Lipid tail beads are polymerised into “rigid” cylinders, of radius r, that span the membranes in a circle of radius Rp An external force, of magnitude Fext, is applied to pull the barrels apart radially MEMPHYS 25
- 26. Proteins in Fusion Transmembrane proteins can exert forces on the bilayer (McNew et al., J. Cell. Biol. 150:105 (2000)) See also Venturoli et al, Biophys. J. 88:1778 (2005) MEMPHYS 26
- 27. Protein-Induced Fusion Protocol Define 6 barrels per membrane: e.g., r = 1.5 a0, Rp = 6 a0 Specify the external force magnitude and direction Measure the time at which the pore first appears and how large it grows (Fusion time definition: time from when Fext > 0 to when pore diameter is > a few amphiphile diameters) Shillcock and Lipowsky, J. Phys. Cond. Mat. 18: S1191 (2006) MEMPHYS 27
- 28. Typical Fusion Event Box = 100 x 100 x 42 nm3 28,000 BLM amphiphiles 3.2 x 106 beads in total 5887 Vesicle amphiphiles MEMPHYS 28
- 29. Dependence on Force 1 8000 2 7000 6000 3 4 runs per applied force Work 5000 4 Duration between 40 ns and 64 ns Done 4000 Barrels move ~ 8nm (4 x their diameter) /kT 3000 If force is too small, no pore appears 2000 1000 0 214 171 150 External Force /pN per barrel NB. Work done is for all 12 barrels MEMPHYS 29
- 30. Nanoparticles and Endocytosis “Rigid” nanoparticles are constructed by tying beads together with Hookean springs giving a “polymerised” surface whose stiffness can be modulated by varying the spring constant Patches created by changing selected bead interactions Star polymers and PEG-ylated lipids are normal DPD molecules MEMPHYS 30
- 31. Nanoparticles in Bulk Proteins are bulky, “rigid” nanoparticles (NP) with sticky patches. What happens if we place them In bulk water? Here are 18 pentagons (shaped like a protein produced by Shigella bacterium), floating in water; The edge and surfaces of each NP Are hydrophobic. MEMPHYS 31
- 32. Nanoparticles near a Membrane What happens if the NPs can interact with a nearby membrane? Here are 9 Shigella proteins floating in water near a fluctuating membrane. The surfaces of each NP are functionalised to adhere to the lipid headgroups, and to aggregate with each other. First, the NPs adhere and slowly diffuse along the surface, next they discover that by aligning in a chain, the membrane can maintain its fluctuations in 1 dimension, and so increase its entropy. MEMPHYS 32
- 33. Nanoparticle Budding How can material pass through a membrane without rupturing it? Some viruses enter a cell by a fusion process that involves them being enveloped in membrane from the target cell. Q What shape of nanoparticle allows it to be enveloped most readily? Here, two rigid nanoparticles are placed near a membrane containing two patches to which the NPs are attracted. The patch lipids are slightly repelled from the surrounding membrane lipids, and the NPs adhere to the patches. The combination of adhesion energy and line tension around the patches drives the budding process. MEMPHYS 33
- 34. Endocytosis How do we construct a coated nanoparticle (NP) in a simulation? (Initial state assembly) NP approaches membrane and cross-links receptors (active binding) Receptors undergo conformational change (modify interactions) NP is internalised in a vesicle (curvature-induction, budding off) NP-vesicle modifies signalling response (???) Experimental questions to answer What selects the NP size and shape that has greatest effect on receptor internalisation? (range is 2 – 100 nm in Jiang et al.) How does the NP surface density of ligands influence receptor response? What influence does the inplane diffusion of receptors have? Nanoparticle-mediated cellular response is size-dependent Jiang et al, Nature Nanotechnology 3:145 (2008)
- 35. Proteins/nanoparticle GNP Size / nm Proteins per Nanoparticle Surface protein density / nm-2
- 36. Polymer-coated nanoparticle Encode self-assembly in polymer’s interactions: H1-[ B B B S6 B B ]-T1 109 comb polymers; hydrophobic backbone and hydrophilic sidechains Spherical nanoparticle with hydrophobic surface Apply forces to arrange the polymers so that they coat the NP MEMPHYS 36
- 37. Coated Nanoparticles We want to make Quantum Dots that consist of a rigid core that is coated by layers of functional polymers: but how do we wrap the core with the polymers? 5 nm diameter core 5 nm diameter core 25 coat molecules 64 coat molecules coat = Comb polymer -(B B B (S) B B )8 - By applying succesive coats we can build up a structured QD MEMPHYS 37
- 38. Nanoparticle Bulk Diffusion 4 polymerised (solid) spheres with 100% hydrophilic surface Box = (25 x 25 x 12.5 nm)3 0.02M HT6 surfactant Spheres diffuse in solvent, as surfactants micellize MEMPHYS 38
- 39. Quantifying Diffusion of Spheres in Bulk Solvent Mean square displacements (MSD) for 4 spheres (R/a0 = 2) in a (32 a0)3 box: averaging over several trajectories gives more accurate results. MEMPHYS 39
- 40. Stokes’ Law R = 2 data from 4 spheres in a 323 box (1 trajectory / 5 cpu-days) R = 4 data from 1 sphere in a 483 box (1 trajectory / 17.5 cpu-days) Fitting R = 4 data from 200-500,000 Fitting the R = 2 data from 200–500,000 and fixing the slope to zero yields: and fixing the slope to zero yields: Intcpt. = 0.0005 +/- 4.10-6 Intcpt. = 0.0011 +/- 2.10-6 We get D = constant / Radius MEMPHYS 40
- 41. Work in progress • Construct a 2 – 100 nm polymer-coated nanoparticle as QD mimic; several layers of coat required – polymer architecture, surface coverage and QD shape are control parameters • Construct a model plasma membrane with diffusing receptors that oligomerize; QDs that can bind to the membrane and occlude receptors; measure signalling pathway • Parallel code to allow 50 nm particles and (500 nm)2 membrane containing receptors, signalling apparatus, … 41
- 42. Conclusions “the limits of your language are the limits of your world” Wittgenstein Computer simulations provide a language for describing dynamical complex systems with (almost) unlimited control DPD captures processes cheaply (calibration of parameters is time- consuming); experimentally invisible data are accessible on 100 nm/10 µs time-scales: parallel code can reach 1 µm and milliseconds. We can observe molecular rearrangements during cellular processes, e.g., fusion, endocytsosis,…; we can test hypotheses about interactions and function; build toy models and compare their predictions to experimental systems; all more cheaply than in a wet lab. 42