Transcription b.pharm
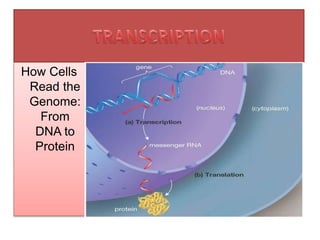
- 1. How Cells Read the Genome: From DNA to Protein
- 2. Transcription • Open DNA at promoter • Make RNA – 5’-> 3’ – transcription bubble • Moves along gene – Prevent DNA knotting • DNA topo-isomerases
- 3. Differences between DNA and RNA RNA DNA 1. Mainly seen in cytoplasm Mostly inside nucleus 2. Usually 100-5000 bases Millions of base pairs 3. Generally single stranded Double stranded 4. Sugar is ribose Sugar is deoxyribose 5. Purines: Adenine, guanine Pyrimidines : cytosine, uracil Adenine, guanine Cytosine, thymine 6. Guanine content is not equal to cytosine and adenine is not equal to uracil Guanine is equal to cytosine and adenine id equal to thymine 7. Easily destroyed by alkali Alkali resistant
- 4. Principal Types of RNAs Produced in Cells
- 5. Introduction Definition: • Transcription is a process in which ribonucleic acid (RNA) is synthesized from DNA. Site: • In prokaryotic cells: ill-defined nuclear zone called the nucleoid whereas In eukaryotic cells : well defined nucleus (transcription of nuclear DNA) or mitochondria (transcription of mitochondrial DNA). Template and Coding Strands: • In transcription, one of the two strands of DNA serves as a template (non- coding strand or antisense strand) and produces working copies of RNA molecules. • The other DNA strand which does not participate in transcription is referred to as coding strand or sense strand or non-template strand. (Coding strand commonly used since with the exception of T for U, primary mRNA contains codons
- 6. Transcription in Prokaryotes • Polymerization catalyzed by RNA polymerase – Can initiate synthesis – Uses NTPs – Requires a template – Unwinds and rewinds DNA • 4 stages – Recognition and binding – Initiation – Elongation – Termination and release
- 7. Transcription is selective • The entire molecule of DNA is not expressed in transcription. • Transcription can take place at any time but only certain selected regions of the DNA are copied. This is like taking Xerox copy of particular page of the book. • So, the genetic information in DNA is transcribed (copied) to the messenger RNA (mRNA).
- 8. SIMILARITY AND DIFFERENTIATION BETWEEN REPLICATION AND TRANSCRIPTION Similarity: 1. They involve the general steps initiation, elongation and termination. 2. Synthesis occurs in the 5’ → 3’ direction. 3. Follows Watson-Crick base pairing rules. Differentiation: 1. Ribonucleotides are used in RNA synthesis rather than deoxyribonucleotides. 2. Uracil replaces thymine as the complementary base pair for adenine in RNA synthesis. 3. A primer is not required in RNA synthesis. 4. Only a small portion of the genome is transcribed into RNA, whereas the entire genome must be copied during DNA replication. 5. RNA polymerase lacks proofreading function during RNA transcription. 6. A single strand of DNA acts as a template for synthesis of particular RNA molecules.
- 9. BASIC REQUIREMENTS FOR RNA SYNTHESIS: 1. DNA template • A single strand of DNA acts as a template to direct the formation of complementary RNA transcript. • The strand that is transcribed to RNA molecule is referred to as the template strand of the DNA. • The other strand is referred to as the coding strand of the gene. 2. Substrate • The substrates for RNA synthesis are the four ribonucleotide triphosphates: I. rATP II. rGTP III. rCTP IV. rUTP 3. Enzyme • DNA dependent RNA polymerase, called RNA polymerase (RNAP), is responsible for the synthesis of RNA, 5’→3’ direction, using DNA template.
- 10. RNA Polymerase • Prokaryotes have single RNA polymerase (RNAP) that transcribes all three RNAs, i.e. mRNA, rRNA and tRNA. • RNA polymerase requires Mg2+ as well as Zn2+ for its activity • 5 subunits, 449 kd (~1/2 size of DNA pol III) • Core enzyme – 2 subunits---hold enzyme together – --- links nucleotides together – ’---binds templates • ---recognition • Holoenzyme= Core + sigma
- 11. ‘holoenzyme’ β β’α2 KD ~ 10-9 M β β’α2 + ‘core’ } Can begin transcription on promoters and can elongate } Can elongate but cannot begin transcription at promoters σ factor is required for bacterial RNA polymerase to initiate transcription on promoters The discovery of initiation factors σ σ
- 12. Template and Coding Strands 5’–TCAGCTCGCTGCTAATGGCC–3’ 3’–AGTCGAGCGACGATTACCGG–5’ 5’–UCAGCUCGCUGCUAAUGGCC–3’ Sense (+) strand DNA coding strand Non-template strand DNA template strand antisense (-) strand RNA transcript transcription
- 13. STAGES OF TRANSCRIPTION • The RNA synthesis involves: 1. Recognition 2. Initiation 3. Elongation 4. Termination
- 14. Recognition • Template strand • Coding strand • Promoters – Binding sites for RNA pol on template strand – ~40 bp of specific sequences with a specific order and distance between them. • Core promoter elements for E. coli – -10 box (Pribnow box) – -35 box • Numbers refer to distance from transcription start site
- 15. Typical Prokaryote Promoter • Pribnow box located at –10 (6-7bp) • -35 sequence ~(6bp) • Consensus sequences: Strongest promoters match consensus – Up mutation: mutation that makes promoter more like consensus – Down Mutation: virtually any mutation that alters a match with the consensus Consensus sequences
- 16. Initiation of RNA chains Binding of RNA polymerase holoenzyme to a promoter region in DNA ( promoter region). Localized unwinding of about 10 nucleotide pairs of the two strands of DNA by RNA polymerase to provide a single-stranded template. Formation of phosphodiester bonds between the first few ribonucleotides in the nascent RNA chain. A purine ribonucleotide (GTP or ATP) is usually the first to be polymerized into the RNA molecule.
- 17. A Typical E. coli Promoter © John Wiley & Sons, Inc. ..,-2,-1,+1,+2,..
- 18. Elongation • By the time 10 nucleotide have been added, Sigma factor is released. • Re- and Un-winding activities results in supercoils. The problem of supercoils is overcome by topoisomerases. • -- RNA polymerase walk (literally) on the DNA 5’ to 3’. • -- RNA polymerase utilizes ribonucleotide triphosphates (ATP, GTP, CTP and UTP) for growing RNA chain. • RNA polymerase binds both DNA template and growing RNA chain.
- 19. Termination Signals in E. coli • Rho-dependent terminators—require a protein factor () • Rho-independent terminators—do not require
- 20. Rho-independent terminators—do not require intrinsic termination)
- 21. • RNA transcription stops • --when the newly synthesized RNA molecule forms a G-C- rich hairpin loop followed by a run of As. • --Create a mechanical stress • --Pulls the poly-U transcript out of the active site of the RNA polymerase. • --A-U has very weak interaction
- 22. Rho-dependent terminators—require extrinsic termination) • Rho-dependent terminators (non-intrinsic) —require a protein factor () and rut site • Rut proteins bind specific RNA sequences (>>Cs and <<<Gs) • ρ factor, binds to the growing RNA (and not to RNA polymerase) or weakly to DNA • In the bound state it acts as ATPase and terminates transcription and releases RNA. • Also responsible for the dissociation of RNA polymerase from DNA Rho utilization (rut)
- 23. Quick review • RNA synthesis occurs in three stages: (1) initiation, (2) elongation, and (3) termination. • RNA polymerases—the enzymes that catalyze transcription—are complex multimeric proteins. • The covalent extension of RNA chains occurs within locally unwound segments of DNA. • Chain elongation stops when RNA polymerase encounters a transcription-termination signal. • Transcription, translocation, and degradation of mRNA molecules often occur simultaneously in prokaryotes.
- 25. Transcription in eukaryotes Introduction: • More complicated process than transcription in prokaryotes. • Three different polymerases. • Each polymerase recognizes a distinct promoter. • Involves separate polymerase for the synthesis of rRNA, tRNA and mRNA. • Eukaryotic RNA polymerase (RNAP) does not include a removable sigma (σ) factor instead, a number of accessory proteins identify promoters and recruit RNAP to the transcription start site.
- 26. Eukaryotic RNA Polymerase RNA Pol. Location Products Alpha- Amanitin Promoter I Nucleolus Large rRNAs (28S, 18S, 5.8S) Insensitive Bipartite promoter II Nucleus Pre-mRNA, some snRNAs Highly sensitive Upstream III Nucleus tRNA, small rRNA (5S), snRNA Intermediate sensitivity Internal promoter and terminator
- 27. Promoter sites • A sequence of DNA bases identical to pribnow box of prokaryotes is identified. • This sequence (TATAAA), known as Hogenes box (or TATA box), is located on the left about 25 nucleotides away (upstream) from the starting site of mRNA synthesis. • Also exists another site of recognition known as CAAT box (GGCCAATCT); between 70 and 80 nucleotides upstream from the start of transcription. • One of these two sites helps RNA polymerase II to recognize the requisite sequence on DNA for transcription.
- 28. Enhancers • Regulate gene expression. • Can increase gene expression by about 100 fold. • Depending upon whether they increase or decrease the initiation rate of transcription, they are called enhancers or repressors. • It is believed that the chromatin forms a loop that allows the promoter and enhancer to be close together in space to facilitate transcription.
- 29. PROCESS TRANSCRIPTION • The process of transcription by RNAP II can be described in terms of following phases. 1. Assembly 2. Initiation 3. Elongation 4. Termination
- 30. ASSEMBLY • The TATA box is the major assembly point for the proteins of the preinitiation complexes of RNAP- II. • The DNA is unwound at the initiator sequence (Inr) and the transcription start site is present within or very near this sequence.
- 31. Initiation of transcription in eukaryotes • First, the TATA box is recognized by TBP (TATA binding protein). • Instead of the sigma factor, SL1 factor ensures that RNAP locates the start point. • In humans about 105 transcription initiation sites are available. • The sequential assembly of TATA binding protein (TBP) bound to TFII A and transcription factors, TFII B, TFII F plus RNAP II, TFII E and TFII H results in a closed complex. • Within the complex the DNA is unwound at the initiator (Inr) region by the helicase activity of TFII H and TFII F, creating an open complex. • The carboxyl terminal domain (CTD) of the largest subunit of RNAPII is phosphorylated by TFII H, and the RNAP II then escapes the promoter and begins transcription.
- 33. Elongation of transcription in eukaryotes • Elongation is accompanied by the release of many transcription factors and is also enhanced by elongation factors. • The function of all elongation factors is to suppress the pausing or arrest of transcription by the RNAPII-TFIIF complex.
- 34. Termination of transcription in eukaryotes • Once the RNA transcript is completed, transcription is terminated RNAPII is dephosphorylated and recycled, ready to initiate another transcript. • The primary mRNA transcript produced by RNA polymerase II in eukaryotes is often referred to as heterogeneous nuclear RNA (hnRNA). • This is then processed to produce mRNA needed for protein synthesis.
- 36. POST-TRANSCRIPTIONAL MODIFICATIONS • The RNAs produced during transcription are called primary transcripts. • They undergo many alterations – terminal base additions, base modifications, splicing etc., which are collectively referred to as post- transcriptional modifications. • This process is required to convert the RNAs into the active forms. • A group of enzymes, namely ribonucleases, are responsible for the processing of tRNAs and rRNAs of both prokaryotes and eukaryotes. SIGNIFICANCE OF POST- TRANSCRIPTIONAL MODIFICATION • Post transcriptional modification of RNA is required for 1. Increased stability of RNA and 2. Regulation of gene expression 3. To convert RNAs into active forms
- 37. PROCESSING OF mRNA • Post-transcriptional modifications of mRNA occur in the nucleus. • Precursor form of mRNA is called as heterogeneous nuclear RNA (hnRNA). • Processing of mRNA from hnRNA includes: 1. 5’ Capping • The 5’ end of mRNA is capped with 7-methylguanosine by an unusual 5’→ 5’ triphosphate linkage. • S-Adenosyl methionine is the donor of methyl group. • This cap is required for increased stability of mRNA due to protection against digestion of ribonucleosome by exonuclease. 2. Poly-A tail • Tailing is addition of 20-250 residues of adenylate (poly-A tail) at the 3’ end. • The reaction is catalyzed by polyadenylate polymerase. • It is added to stabilize mRNA. 3. Removal of extra RNA at 3’ end • There are extra-nucleotides at 3’ end of mRNA in the primary transcript. • These extra-nucleotides are cleaved by ribonuclease at 3’ end.
- 38. 4. Splicing • Introns are the intervening nucleotide sequences in mRNA which do not code for proteins. • Exons of mRNA possesses genetic code and are responsible for protein synthesis. • The removal of introns is promoted by small nuclear ribonucleoprotein particles (snRNA). • The spliceosome is used to represent the snRNP association with hnRNA at the exon- intron junction. • The processing of hnRNA molecules becomes a site for the regulation of gene expression. • The mature RNA then enters the cytosol to perform to perform its function (translation). Faulty splicing may result in diseases. • A good example is one type of β-thalassemia. • β-thalassemia is a genetic disease characterized by impaired synthesis of β-globin protein. • This leads to impaired production of hemoglobin and anemia. • The β globin gene has three exons and two introns. • The introns contain a G-T sequence that is essential for the correct splicing of the RNA and the formation of normal mature β globin mRNA. • In β-thalassemia, the G-T sequence is mutated to A-T sequence resulting in defective RNA splicing. • The defective β globin mRNA cannot be utilized for translation.
- 40. PROCESSING OF PRECURSOR tRNA • The four different events of post-transcriptional processing of pre-tRNA molecules are 1. Removal of extra RNA • These are extra nucleotides at 5’ end and 3’ end of tRNA in the primary transcript. These extra nucleotides are cleaved by ribonuclease P at the 5’ end and ribonuclease D homologue in eukaryotes at 3’ end. 2. Removal of Introns from Anticodon Site • Some pre tRNA molecules may have a short introns at the anticodon site. • These are removed by splicing but mechanisms are different from pre mRNA splicing. 3. Addition of CCA sequence • The sequence CCA is added to the 3’ end of all the t RNAs. • The free OH group present at the 3’ end is required for the formation of amino acyl t RNA during protein synthesis. 4. Modifications of Bases • Several nucleotides in pre t RNA molecules undergo modification. • These include formation of unusual bases such as pseudo-uridine, dihydrouracil, thymine and methylated bases.
- 41. PROCESSING OF PRE-RIBOSOMAL RNA 1. 45 S Precursor RNA • Most of the eukaryote have more than 100 copies of rRNA genes because of the requirement for large number of ribosomes (containing rRNA) for protein synthesis. • The genes of these three RNA are thus clustered together and tandemly repeated with spacer sequences in between them. • The cleavage of the 45 S rRNA results in the release of 18S, 5.8 S and 28 S RNAs. • Methylation of bases occurs before cleavage. 2. 5S RNA • The 5S RNA is also present in multiple copies. • 5s rRNA migrate to the nucleolus and does not undergo any processing.
- 43. INHIBITORS OF TRANSCRIPTION • The synthesis of RNA is inhibited by certain antibiotics and toxins. 1. Rifampicin • Rifampicin and streptovaricin bind with β subunit of the polymerase to block the initiation of transcription. • It is an antibiotic widely used for the treatment of tuberculosis and leprosy. 2. Actinomycin D • This is also known as dactinomycin. • It forms a complex with double stranded DNA and prevents the movement of core enzyme and as a result inhibits the process of chain elongation. • This was the first antibiotic used for the treatment of tumors. 3. Streptoglydigin • It binds with the β subunit of prokaryotic polymerase and thus inhibits the elongation. 4. Heparin • It is a polyanion that binds to the β’ subunit and inhibits transcription. 5. α-amanitin • It is a toxin produced by mushroom, Amanita phalloides. • Amanitin strongly inhibits RNA polymerase II, inhibiting specifically the biosynthesis of mRNA. 6. Daunomycin • It is an anthracyclin glycoside. • The anthracyclin ring of daunomycin intercalates with double stranded DNA. • It is used in the treatment of cancer.
- 44. REVERSE TRANSCRIPTION DEFINITION • Reverse transcription is the formation of DNA from RNA. • The enzyme responsible for the reverse transcription is called reverse transcriptase. • It is a RNA dependent DNA polymerase. REVERSE TRANSCRIPTION PROCESS • In reverse transcription, the RNA is used to synthesize a new strand of DNA by viral encoded reverse transcriptase (also called RNA dependent DNA polymerase). • The RNA component from the RNA-DNA hybrid is then hydrolyzed by a specific RNA hydrolyzing enzyme, RNAase H. • The DNA strand remained after the hydrolysis of RNA, is used as a template to generate double stranded DNA.
- 47. SIGNIFICANCE 1. Tumor viruses, proto-oncogenes and oncogenes and development of cancer • A number of viruses contain genome containing an RNA molecule instead of DNA. • Examples are retroviruses such as HIV. • The copy of DNA from viral DNA may be incorporated to host DNA. • Thus, a new virus proteins are produced which can alter cellular function including the malignant transformation of the cell (tumor production). • Examples are protooncongenes and oncogenes derived from retroviruses. 2. Reverse transcription in genetic recombination through transposition • Transposition refers to jumping of mobile DNA sequences (also called transposable elements or transposons) from one place in the genome to another place in the genome. • One of the mechanisms of transposition involves transposable elements jumping through the formation of RNA intermediates. • Transposable elements replicate and move to other genomic sites by transcription to RNA and subsequent reverse transcription of the RNA into a double stranded DNA.
- 48. 3. Telomere replication • The presence of telomere at the end of chromosome allows replication of DNA to its full length. • Impairment of telomere replication leads to loss of genetic information and contributes to aging process. • Reactivation of telomerase activity results in cell proliferation (cancer). 4. Uses of reverse transcriptase in molecular biotechnology • Reverse transcriptase derived from RNA tumor viruses is used to synthesize complementary DNA (cDNA) copies of mRNA templates. • cDNA is used for cloning or reverse transcription-polymerase chain reaction.
