EVE161 Lecture 3
- 1. Lecture 3: EVE 161: Microbial Phylogenomics ! Lecture #3: Era I: Woese and the Tree of Life ! UC Davis, Winter 2014 Instructor: Jonathan Eisen Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 2. Where we are going and where we have been • Previous lecture: ! 2. Evolution of DNA sequencing • Current Lecture: ! 3. Woese and the Tree of Life • Next Lecture: ! 4. Modern view of Tree of Life Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 3. Era I: rRNA Tree of Life Era I: rRNA Tree of Life Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !3
- 4. • Tree of life vs. Trees of Life Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !4
- 5. Phylogeny was central to Darwin’s Work on Natural Selection Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !5
- 6. Phylogeny • Phylogeny is a description of the evolutionary history of relationships among organisms (or their parts). • This is portrayed in a diagram called a phylogenetic tree. • Phylogenetic trees are used to depict the evolutionary history of populations, species and genes. • The Tree of Life refers to the concept that all living organisms are related to one another through shared ancestry. Ch. 25.1 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !6
- 7. Darwin and a Single Tree of Life George Richmond. Darwin Heirlooms Trust Darwin Origin of Species 1859 ! Set stage for “tree thinking” Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !7
- 8. How many trees of tree The default class life? If this tree included all major groups of organisms a b c d e h g f What does this imply about the # of times life evolved? Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !8
- 9. Common ancestry of all life a b c d e h g f MRCA Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !9
- 10. Common ancestry of all life a b c This implies a single origin of life, sometime before MRCA d e h g f MRCA Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !10
- 11. Common ancestry of all life a b c This implies a single origin of life, sometime before MRCA d e h MRCA g f And thus that there is one “Tree of Life” Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !11
- 12. What if life originated twice? But no theoretical reason why we could living organisms could not have multiple separate ancestries a b c d e h g f Origin 1 Origin 2 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !12
- 13. How many trees of life? • How do we distinguish two hypotheses? ! 1. a single origin (and thus a single tree of life) 2. multiple origins (and thus multiple separate trees)? ! • What data might support one hypothesis versus the other? Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !13
- 14. Universal homologies • Characters that are found in the same state an all organisms are considered “universal” (remember “presence” can be a state) • Characters (i.e., features like the smiley face) that are inherited from a common ancestor are homologous • Homologous characters that are found in the same state in all organisms are “universal homologies” Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !14
- 15. 12.3 From Gene to Protein Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !15
- 16. 12.6 The Genetic Code Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !16
- 17. The Ribosome Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !17
- 18. Universal homologies: examples • Molecular and cellular features ! Use of DNA as a genetic material ! Use of ACTG in DNA ! Use of ACUG in RNA ! Three letter genetic code ! Central dogma (DNA -> RNA -> protein) ! Lipoprotein cell envelope ! 20 core amino acids in proteins • Specific complexes and genes ! Ribosomal proteins and RNA ! RNA polymerase proteins Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !18
- 19. Universal homologies: examples • Data matrix for universal homologies ! DNA Three letter code DNA-> RNA> protein E. coli + + + Yeast + + + Humans + + + M. jannascii + + + Organism • What does this mean in terms of the number of “origins” of life? Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !19
- 20. How to infer a phylogenetic tree Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !20
- 21. What if life originated twice? a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !21
- 22. What if life originated twice? a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !22
- 23. What if life originated twice? a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !23
- 24. What if life originated twice? a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !24
- 25. What if life originated twice? a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !25
- 26. a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !26
- 27. a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !27
- 28. a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !28
- 29. a b c d e h g Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 f !29
- 30. a b c d e h g f Problem: homologous trait needs to have a single origin Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !30
- 31. a b c d e h g f Must connect the trees Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !31
- 32. a b c d e h g f Must connect the trees Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !32
- 33. a b c d e h g Conclusion: existence of universal homologies Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 implies a single tree of life Slides for UC f !33
- 34. a b c d e h g f MRCA Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !34
- 35. Universal homologies • What does the existence of “universal homologies” mean? • The key to the implications of “universal homologies” to the Tree of Life, is that homologous traits should be derived from a single, common ancestral trait. • This implies a single origin of life. • Note - lateral transfer can lead to organisms sharing homologous features EVEN if the organisms have a separate origin Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !35
- 36. LUCA Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !36
- 37. How to Build a Tree of Life? Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !37
- 38. • Data matrix for universal homologies ! DNA Three letter code DNA-> RNA> protein E. coli + + + Yeast + + + Humans + + + M. jannascii + + + Organism • What does this mean in terms of the relationships among different branches? Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !38
- 39. • For building a single tree including all organisms • Need traits that are homologous between all organisms but which have variable states • Examples??? ! Until 1960s these were hard to come by ! Researchers focused instead on relationships within particular branches of the Tree of Life ! Within sub-branches, more possible traits to use (e.g., bones in mammals) Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !39
- 40. Ernst Haeckel 1866 Plantae Protista Animalia Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !40
- 41. Whittaker – Five Kingdoms 1969 Monera Protista Plantae Fungi Animalia Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !41
- 42. Carl Woese http://mcb.illinois.edu/faculty/ profile/1204 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !42
- 43. Two papers for today Proc. Natl. Acad. Sci. USA Vol. 74, No. 10, pp. 4537-4541, October 1977 Evolution Classification of methanogenic bacteria by 16S ribosomal RNA characterization (comparative oligonucleotide cataloging/phylogeny/molecular evolution) GEORGE E. Fox* t, LINDA J. MAGRUM*, WILLIAM E. BALCHt, RALPH S. WOLFEf, AND CARL R. WOESE** Departments of *Genetics and Development and tMicrobiology, University of Illinois, Urbana, Illinois 61801 Communicated by H. A. Barker, August 10, 1977 residues represented by hexamers and larger in catalog A and in catalog B and their overlap of common sequences, respectively. The association coefficient, SAB, SO defined provides what is generally an underestimate of the true degree of homology between two catalogs because related but nonidentical oligomers are not considered. The matrix of SAB values for each binary comparison among the members of a given set of organisms is used to generate a dendrogram by average linkage (between the merged groups) clustering. The resulting denmetabolic capacity to growstructure oxidizing hydroanaerobically by of the prokaryotic strictly speaking, phyletic because no "ancestral drogram is, domain: The primary Phylogenetic catalog" has been postulated. However, it is clear from the gen and reducing carbon dioxide to methane (1-3). Their rekingdoms molecular nature of the data that the topology of this dendrolationships to one another and to other microbes remain virunknown. Protein and nucleic acid primary structures gram would closely resemble, if not be identical to, that of a tually(archaebacteria/eubacteria/urkaryote/16S ribosomal RNA/molecular phylogeny) are perhaps the most reliable indicators of phylogenetic relaphylogenetic tree based upon such ancestral catalogs. tionships R. WOESE AND GEORGE E.as the 16S ribosomal CARL (4-6). By using a molecule, such Fox* constrained RNA, that is readily isolated, ubiquitous, and highly RESULTS inDepartment of Geneticspossible to relateUniversity ofmost distant of sequence (7), it is and Development, even the Illinois, Urbana, Illinois 61801 The 10 organisms whose 16S ribosomal RNA oligonucleotide microbial species. To date, approximately 60 bacterial species Communicated by T. M. Sonneborn, August16S ribosomal RNA 18,1977 catalogs are listed in Tables 1 and 2 cover all of the major types have been characterized in terms of their of methanogens now in pure culture except for 2; we have been primary structures (refs. 6-9, unpublished data). We present A phylogenetic analysis based upon by this to construct a culture of Methanococcus between domains: ABSTRACT here results of a comparative study of the methanogensribosomal unable to obtain phylogenetic classificationsvanniehfi (19), and RNA sequence characterization reveals one another and to that living systems Methanobacterium mobile (20) comparabledifficult to grow Prokaryotic kingdoms are not has proven to eukaryotic ones. method, which shows their relationships to represent one of This should sequences in by, an bear little terminology. The typical bacteria. of three aboriginal lines (ii) descent: (i) the eu- and label. The be recognizedTable 1appropriateresemblance to bacteria, comprising all typical bacteria; the archaebacteria, those for typical bacteria (refs. the prokaryotic domain we think 6-9; unpublished data). Fig. 1 highest containing methanogenic bacteria; and (iii) the urkaryotes, now is ashould phylogenetic unit inthe SAB values in Table 3."primary dendrogram derived from called an "urkingdom"-or perhaps It METHODScomponent EVE161 Courseseen be by Jonathan Eisen Winter 2014 can Slides for UC Taught methanogens comprise two major divisions. The represented in the cytoplasmic Davis of eukaryotic be kingdom." This would recognize the qualitative distinction that-the cells. ABSTRACT The 16S ribosomal RNAs from 10 species of methanogenic bacteria have been characterized in terms of the oligonucleotides produced by T1 RNase digestion. Comparative analysis of these data reveals the methanogens to constitute a distinct -phylogenetic group containing two major divisions. These Natl. Acad. Sci. USAto be only distantly related to typical Proc. organisms appear bacteria.No. 11, pp. 5088-5090, November 1977 Vol. 74, Evolution The methane-producing bacteria are a poorly studied collection of morphologically diverse organisms that share the common !43
- 44. Two papers for today Proc. Natl. Acad. Sci. USA Vol. 74, No. 10, pp. 4537-4541, October 1977 Evolution Classification of methanogenic bacteria by 16S ribosomal RNA characterization (comparative oligonucleotide cataloging/phylogeny/molecular evolution) GEORGE E. Fox* t, LINDA J. MAGRUM*, WILLIAM E. BALCHt, RALPH S. WOLFEf, AND CARL R. WOESE** Departments of *Genetics and Development and tMicrobiology, University of Illinois, Urbana, Illinois 61801 Communicated by H. A. Barker, August 10, 1977 ABSTRACT The 16S ribosomal RNAs from 10 species of methanogenic bacteria have been characterized in terms of the oligonucleotides produced by T1 RNase digestion. Comparative analysis of these data reveals the methanogens to constitute a distinct -phylogenetic group containing two major divisions. These organisms appear to be only distantly related to typical bacteria. residues represented by hexamers and larger in catalog A and in catalog B and their overlap of common sequences, respectively. The association coefficient, SAB, SO defined provides what is generally an underestimate of the true degree of homology between two catalogs because related but nonidentical oligomers are not considered. The matrix of SAB values for each binary comparison among the members of a given set of organisms is used to generate a dendrogram by average linkage (between the merged groups) clustering. The resulting dendrogram is, strictly speaking, phyletic because no "ancestral catalog" has been postulated. However, it is clear from the molecular nature of the data that the topology of this dendrogram would closely resemble, if not be identical to, that of a phylogenetic tree based upon such ancestral catalogs. The methane-producing bacteria are a poorly studied collection of morphologically diverse organisms that share the common metabolic capacity to grow anaerobically by oxidizing hydrogen and reducing carbon dioxide to methane (1-3). Their relationships to one another and to other microbes remain virtually unknown. Protein and nucleic acid primary structures are perhaps the most reliable indicators of phylogenetic relaSlides for UC Davis the 16S Course Taught by Jonathan Eisen Winter 2014 tionships (4-6). By using a molecule, such asEVE161 ribosomal !44
- 45. • ABSTRACT: ! • The 16S ribosomal RNAs from 10 species of methanogenic bacteria have been characterized in terms of the oligonucleotides produced by T(1) RNase digestion. Comparative analysis of these data reveals the methanogens to constitute a distinct phylogenetic group containing two major divisions. These organisms appear to be only distantly related to typical bacteria. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !45
- 46. 4538 Proc. Nati.'Acad. Sci. USA 74 (1977) Evolution: Fox et al. Oligonucleotide sequence 5-mers CCCCG Table 1. Oligonucleotide catalogs for 16S rRNA of 10 methanogens Present in Present in OligonuOligonucleotide cleotide organism organism number number sequence sequence 1-lO;l,5,8 CCCAG CCACG ACCCG CCAAG CACAG CAACG ACACG ACCAG AACCG ACAAG AAACG AAAAG 6 10 10 9 9 CUCCG CCCUG UCCAG CUCAG OCAUG UCACG UACCG 4,7 ACCUG ACUCG AUCCG UAACG CAAUG ACUAG ACAUG AUACG AAUCG UAAAG AUAAG AAAUG UUCCG CUUCG UCCUG CCUUG CUCUG UCUAG UUCAG CUAUG UACUG UAUCG ACUUG AUCUG AUUCG UUAAG UAAUG AUAUG AAUUG AUUAG UUUCG UUCUG UCUUG CUUUG UUUAG UUAUG UUUUG 6-mers CCCCAG 1-10;8-9 7-9 7 1-10;10 1-6;1,5 7-9 1,6,9-10 9 6-8s1 1-10 1-10 1-2,4-5 1-6,8 4-5;5 6 9 4-9 1-6;4 2-3,8-9 10 7 10 2 3-10;3,6-9;7 4 1-6,8;4 5-6,8 1-6;4 1 6,8 7 5,7-9;9 5 7-10;8-10 7-8 1-6,10 3-5,7-8 2-3,10 1-10;1-2,4,6,8,10 1-2,5,10;2 3-4,9 1-10;1-2,4-6,9 CCCUUG CCUCUG UCCCUG CCUUAG CUCUAG CUUCAG UCCUAG UUCCAG CCUAUG CUACUG UCACUG CUAUCG UCAUCG CAUCUG ACUCUG ACCUUG AUCCUG UCUAAG UUACAG UAUCAG UAUACG UAAUCG AUACUG ACAUUG AACUUG AAUCUG UAAAUG AUUAAG AAUAUG 4 1 4-5,7-9 1-6,10;1 7,9-10 .7-9 1-6,8-10 9 1-10 10 1-6,10 10;10 1-10;8--9 1-3,7 1-2,4-6 6,10 7-9 1-10 4,7-8 1-3 9 1-2 1-6 3 1-3,6 3,7-9 7-10 4,60 7-9 6-10 8-10 CCCUCG CCUCAG CUCCAG UCCCAG CCACUG ACCUCG CCUAAG CUCAAG 5,8,10 5 AACCCCG CCAACAG CAACACG CAAACCG CCCUACG CCCACUG SUCCACCG CCACCUG 2,7-10 CCCUAAG 4-5,9 9 UCACACG CUACACG 1-3,5,10 4-6 UAACCCG 7 6 3 6 10 7-9 10 1 1-2,4,7-8 1-2,5 5UAUUUUG UUCUUUG 4-6 3 1-4,6 UUUUUUG 1-3 2,8-10 8-mers CCACAACG ACCCCAAG AAACCCCG 9 7 6,10 1-2,4-6,10 3-4 -1-4,7-8,10 1-2,4-6 UUUUUG ACCACG ACACCG AAACCG 7-10 7 8 9 7 10 7-8 10 1 1 10 1 3 5-9 4 1-8 9 2,9 CCCCAG 1,3-6 1,3-6 UUCUUCG UCUCUUG CUUUAUG UUUAUCG UAUUUCG AUUAUUG 3,7-8,10 1-5;1 1,4 ~~~~~ACCCACG ACCACCG 10 7-9 9 7 2 8 10 10 4 3 CUUUUG UCUUUG 7-mers AAAUCUG 8-1o CUCCUUG UCCCUUG UUCUCCG CUCUUAG UACUUCG UACUCUG UCAUAUG UAAUCUG AAUUUAG 1-3,5,10 2,7 4,9;9 4,7,9 CCCUUAG CAUCCUG UACUCCG AUCUCCG ACCUUCG UCCUAAG UUACCAG CUAACUG UAACUCG AUUCCAG AUCAUCG AAUCUCG AACCUUG UCUAAAG CUUAAAG CAAUAUG AUACUAG AAUCUAG number 1-10 8 1-6 1-10 7 10 10 7 7-8 4-6 1-10 7-8 8 9 7 1-10 3 8-9 1-10;1l-7,9;7 AUACCCG AACCUCG CCUAAAG UAACACG AUAACCG AAUCCAG AACAUCG AAAUCCG UAAAAAG organism UAAAAUG 7,9 CCUUUG CUUUCG UCUCUG UUCCUG UCUUAG CUAUUG UUACUG UAUUCG AUUCUG ACUUUG UAUAUG 4,6 CACCAG CCAUAG CAUACG ACACUG AACCUG AAUCCG CUAAAG UAAACG ACUAAG ACAAUG AUAACG AAUACG AACAUG AAACUG AAAUCG AAUAAG Present in 10 10 3 2 8 1-9 7 7 UCCACCAG CCCACAUG CUCAACCG ACCCUCAG ACCACCUG UAACACCG AUCCCAAG AAAUCCCG 1-2,5-6 CCCUCAUG UACUCCCG 1-10 10 CCUAUCAG CCUAACUG CUUAACCG 7-8 8-9 4-6 10 7-8 3 4,7-10 5-6 AU(CCUC)CG TUAAUCCCG CUACAAUG UACUACAG UAAUACCG AUUACCAG AUAACCUG 1-3,5-10 1-3,5-6,9-10 1,5 9 7-8 8 7 1,3-6,8,10 1-6,10 2-3 1 1,3-4 4 5 10 5 4,7,9 9 1-10 10 7-9 3 6-8,10 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 Table 1 continues on following page. !46
- 47. Proc. Nati. Acad. Sci. USA 74 (1977) Evolution: Fox et al. 4539 Table 1. (continued) Oligonu- Present in Oligonu- Present in Oligonu- Present in sequence number sequence number sequence number cleotide organism cleotide organism ACAAUCUG AAAUCCUG AUAAACUG AUAAAUAG 9 1-2,6-9 AAUUAUCCG UUUAAAACG UAAACUAUG AUAAUACUG (CU,CCUU)CG 4 4 CUAUUACUG UUAAAUUCG UUUAAUAAG 9 1 2 UUAUAUUCG UAUUUCUAG UUUAUUAAG 2 9 CUUUUAUUG 6 AUCCUUCG UCUAACUG CUUAACUG UAAUCCUG UCUAAAUG UUAAAUCG CAUAUAUG AAAUCUUG AAAUUCUG AUAAAUUG 3-6 2 1 2-3,5-6 1-3,6 1 10 10 10 2-3 1 7-9 7 7 2 1 cleotide organism UUUUUUUCCUG UUUUUUUUAAG 1 2 12-mers CCACCCAAAAAG UCAAACCACCCG UCAAACCAUCCG ACAUCUCACCAG CCACUCUUAACG CCAUUCUUAACG CUCAACUAUUAG CCACUAUUAUUG CAAUUAUUCCUG CCACUUUUAUUG CCAUUUUUAUUG UUUUUAUUG 2,4 (CUA,CUUUUA)UUG UUUUUUUCG 1 CUUUUCAG 613mr 2 UUCUCAUG 10-mers UAAACUACACCUG UUUAAUCG 9 7 AAUAACCCCG (CAA,CCA)CAUUCUG UAUCAUUG 9 2-3 UUUAAAUG ACCACCUAUG 9UCAAAC 8 AAUCUCACCG AUAAUUUUUCCUG UUUAAUUG 1-8,10 AAAUCUCACG 4(UUU,CUU,CU)AAAUG 8 UAACUCAAAG UUUUUUCG 2-3 AAACUUAAAG 1-10 14-mers UUUUAUUG 1 10 ACCUUACCUG AAAACUUUACAAUG 9-mers UUACCAUCAG 3 AUUUUU(CCU,CU)UUG 4-5 CCCACCAAG 10 UACCUACUAG 1-10 CACACACCG 15mr AAUCACUUCG UCUAACACCCGs 8 (CCA,CAA)CAG 6 AACCCUUAUG CAUAAACCCACCUUG CCCAACAAG 7-9 9 UAAAUAACUG AUAACCUAACCUUAG AACCCCAAG 6 AAACCCAAG 4 6 UUCUUCACCG AAUAAUACCCUAUAG ACUCUACUUG 9AAUAAUACUCCAUAG 1 CCUCACCAG 8 CUUAACUAUG AUAAUCUACCCUUAG CCUACCAAG 6 AUACUAUUAG 2,4-5 CCUACAACG 10 AUAACCCCG 6,8,10 UUCCCUAUUG AAAUCCUAUAAUCCCG 1-6 AAACCUCCG UCUUCUUAAG 4 AAUCUCCUAUAAUCAUG CACACUAAG 1-6 CAAUCUCUUAAACCUAG AUAAACCCG 6 1 AUUUUUUUCG UAAUCUCCUAAACCUG UACUCCCAG UUUUUGAAAUCCUAUAAUCCUG UAAUCCCCG 7 AAUCCCCUG 1,3-6 11-mers 17-mers CUUACCAAG 1-3 ACAACUCACCG 10 CAAUCUUUUAAACCUAG 3 (UC)ACACAUG AAAUCCCACAG 6 UAAU(CCU,CU)AAACUUAG 2-3 (UC)ACAAUCG CAUCUCACCAG 4 UCAUAACCG AUAAU(CCU,CU)AAACCUG 7,9 UAACUCACCCG CUAAUACCG 9 3 AAAUCUCACCG ACCCUUAAG 18-mer 7 7,9 AAACACCUUCG 6 AACAAUCUCCUAAACCUG AUAAUCCCG 9 AAAUCCCAUAG AUAACCCUG 1-5 5 AUAAUACCG 24-mer 4-5 UCCCUCCCCUG 10 AUAUACAAG 9 (AAACA,UAAUCUCA)CAUAUCCUCCG 10 CCCAUCCUUAG AAAUCCUAUAG 3 UCUUACCAG 10 UCACUAUCG 6 termini 5' end UUUCAACAUAG UAAUCCCUG 10 7,9 6 pAG UAAUCCUCG A(UA,UCA,CUA)UG 8 pAAUCCG AAUUUCCCG 10 UUUCAAUAUAG pAAUCUG 10 AAUCCUCUG 2 pAUUCUG UCAUAAUCG 1,5 CUUUUCUUAAG CUAAUACUG 1 1,3 CUUUUCAUUAG CAUCAUAUG 2 10 3'end UUCUUUAAUCG 7 AUAAUUCCG 10 AUCACCUCCUOH 1-2,4,6 8-10 7 1-6 4-6 1-3 10 7 2 8 5 3 10 6 ~~~~~~~~~~~~~~~~~~~~~~~~~~UAAUACUCCAUAG9 3 5 9 ~~~~~~~~~~~~~~~~~~~~~~~~~~~AAAACUUUACCAUG 7-8,10 2 1- 14 8 7 5 U4AUCCCAAACAGs 1-3,5-6UUUUU 4 4 7 4 5 3 1-2 9 8 10 4,6 5 1, 2,7-10 1-10 First column is oligonucleotide sequence; second column shows organisms in which that sequence is found. Organisms are designated by number (see Fig. 1) as follows: 1, M. arbophilicum; 2, M. ruminantium strain PS; 3, M. ruminantium strain M-1; 4, M. formicicum; 5, M. sp. strain M~o.H.; 6, M. thermoautotrophicum; 7, Cariaco isolate JR-i; 8, Black Sea isolate JR-i; 9, Methanospirillum hungatii; 10, Methanosarcina barkeri. Multiple occurrences of a sequence in a given Course Taught by Jonathan Eisen Winter 2014 Slidesa doubleUC Davis EVE161organism are denoted by repeating the organism's number in column 2: e.g., 1-4,6-8;3,7,;3 signifies for occurrence in organism 7 and a triple occurrence in organism 3. !47
- 48. 4540 Evolution: Fox et al. Proc. Nat Table 2. Post-transcriptionally modified sequences and likely counterparts Occurrence in methanogens Occurrence in IA Sequence IB IIA IIB typical bacteria 1.AACCUG + + 30% AAUCUG + + None AAG 55% 2. UAACAAG + + None UAACAAG + + None UAACAAG >95% 3. AUNCAACG + + None ACNCAACG + + None AX6CTAACG - >90% 4. NCCG + + None + + C((CC)G None N'CCG >95% 5. CC(CCG + >95% Post-transciptionally modified sequences in methanogens and their likely counterparts in the bacteria that have been examined. In group 1, A is N-6-diMe (21), identified by electrophoretic mobilities of A and AA and by total resistance to U2 nuclease. In group 2, U is partially resistant to pancreatic nuclease, the first A when modified is still U2 nuclease sensitive; the second A is N-6-diMe. N in group 3 is resistant to pancreatic nuclease but is electrophoretically U-like. X stands for U or A. In group 4, N and N' are not cleaved by endonucle~aes; NC and N'C are electrophoretically distinguishable; C is cleaved by pancreatic nuclease and has C-like electrophoretic prop,erties. In group 5, IC (21, 22) is not cleaved by pancreatic nuclease and is readily deaminated by NH40H. phylogenetic distinction Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 A of this apparent magnitude is nication). The extent t unique remains to be d (iii) All other bacteri single exception of the glycan (26, 27). Howeve examples) do not contai personal communication (iv) Table 2 shows th 16S ribosomal RNA in ferent from that in typic as well (D. Stahl, p methanogens are the fi terized (prokaryote or called "common sequen contain a I"ICG seque U'ICG (the dot above a cation; U T) (L. data). It should be noted th be completely unrelated requirement of a strict become the more impre ogens have been charact biochemistry and molec It would appear that me classified as a systemat (inclusive of the blue-g Although it cannot methanogens represent !48
- 49. ilarly, coenzyme F420, which handles low-potential electrons, is present in all methanogens but so far is not found elsewhere Note Added in Proof: Preliminary characterization of Methanobacterium mobile, a motile, Gram-negative, short rod, places this organism in group IIA. Methanobacterium sp. strain AZ (30) has been shown to be a strain of M. arbophilicum; SAB = 0.87 for the pair. The work reported herein was performed under National Aeronautics and Space Administration Grant NSG-7044 and National (25). (ii) We have been unable to detect cytochromes in these organisms, and R. Thauer obtained no evidence for the presence of quinones in M. thermoautotrophicum (personal commu- Table 3. SAB values for each indicated binary comparison Organism 1. M. arbophilicum 1 2 3 4 5 6 Organism 7 8 9 2. M. ruminantium PS .66 3. M. ruminantium M-1 .60 .60 4. M. formicicum .50 .48 .49 5. M. sp. M.o.H. .53 .49 .51 .60 6. M. thermoautotrophicum .52 .49 .51 .54 .60 7. Cariaco isolate JR-1 .25 .27 .25 .26 .23 .25 8. Black Sea isolate JR-1 .26 .28 .26 .27 .28 .29 .59 9. Methanospirillum hungatii .20 .24 .21 .22 .23 .23 .51 .52 10. Methanosarcina barkeri .24 .41 .29 .26 .24 .26 .34 .25 .33 11. Enteric-vibrio sp. .08 .08 .11 .05 .06 .07 .09 .09 .10 12. Bacillus sp. .10 .14 .11 .11 .12 .10 .08 .10 .10 13. Blue-green sp. .10 .11 .10 .08 .09 .10 .10 .10 .08 The values given for enteric-vibrio sp., Bacillus sp., and blue-green sp. represent averages obtained from 11 species, respectively. 10 11 12 13 .10 .27 .08 .11 .24 .26 (9), 7 (6), and 4 (23) individual Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !49
- 50. 50 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !50
- 51. Two papers for today Proc. Natl. Acad. Sci. USA Vol. 74, No. 11, pp. 5088-5090, November 1977 Evolution Phylogenetic structure of the prokaryotic domain: The primary kingdoms (archaebacteria/eubacteria/urkaryote/16S ribosomal RNA/molecular phylogeny) CARL R. WOESE AND GEORGE E. Fox* Department of Genetics and Development, University of Illinois, Urbana, Illinois 61801 Communicated by T. M. Sonneborn, August 18,1977 to construct phylogenetic classifications between domains Prokaryotic kingdoms are not comparable to eukaryotic ones This should be recognized by, an appropriate terminology. Th highest phylogenetic unit in the prokaryotic domain we thin should be called an urkingdom-or perhaps primar kingdom. This would recognize the qualitative distincti between prokaryotic and eukaryotic kingdoms and emphasiz The biologist has customarily structured his world in terms of that the former have primary evolutionary status. certain basic dichotomies. Classically, what was not plant was The passage from one domain to a higher one then become animal. The discovery that bacteria, which initially had been a central problem. Initially one would like to know whether th considered plants, resembled both plants and animals less than is a frequent or a rare (unique) evolutionary event. It is trad plants and animals resembled one another led to a reformulationally assumed-without evidence-that the eukaryot tion of the issue in terms of a yet more basic dichotomy, that of domain has arisen but once; all extant eukaryotes stem from eukaryote versus prokaryote. The striking differences betweenTaught common ancestor, itself eukaryotic (2). A similar prejudice1 Slides for UC Davis EVE161 Course by Jonathan Eisen Winter 2014 !5 hol A phylogenetic analysis based upon ribosomal ABSTRACT RNA sequence characterization reveals that living systems represent one of three aboriginal lines of descent: (i) the eubacteria, comprising all typical bacteria; (ii) the archaebacteria, containing methanogenic bacteria; and (iii) the urkaryotes, now represented in the cytoplasmic component of eukaryotic cells.
- 52. Woese and Fox • Abstract: A phylogenetic analysis based upon ribosomal RNA sequence characterization reveals that living systems represent one of three aboriginal lines of descent: (i) the eubacteria, comprising all typical bacteria; (ii) the archaebacteria, containing methanogenic bacteria; and (iii) the urkaryotes, now represented in the cytoplasmic component of eukaryotic cells. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !52
- 53. 12.3 From Gene to Protein Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !53
- 54. The Ribosome Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 54 !54
- 55. rRNA structure Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !55
- 56. rRNA Systematics • All cellular organisms have ribosomes • All have homologous subunits of the ribosomes including specific ribosomal proteins and ribosomal RNAs (i.e., these are universally homologous genes) • Woese determined the sequences of ribosomal RNAs from different species • The sequences are highly similar but have some variation • Each position in a rRNA can be considered a distinct character trait • Each position has multiple possible character states (A, C, U, G) Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !56
- 57. Why rRNA for Woese? • • • • Universal Highly conserved functionally Evolves slowly Easy to extract and sequence Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 !57
- 58. Woese Methods • • • • • • Digest rRNA with T1 Rnase 2D electrophoresis Fingerprint Sequence each fragment Oligonucleotide catalog Association constant SAB = 2NAB/(NA+NB) – NA = total number of nucleotides in catalog for A – NB = total number in nucleotides in catalog for B – NAB = total number in the shared catalog Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 59. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 60. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 61. Eubacteria “A comparative analysis of these data, summarized in Table 1, shows that the organisms clearly cluster into several primary kingdoms. The first of these contains all of the typical bacteria so far characterized .... (lots of names here) ... It is appropriate to call this urkingdom the eubacteria.” 61 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 62. Urkaryotes • “A second group is defined by the 18S rRNAs of the eukaryotic cytoplasm-animal, plant, fungal, and slime mold (unpublished data).” • (They call this lineage the urkaryotes) Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 63. Archaebacteria “Eubacteria and urkaryotes correspond approximately to the conventional categories prokaryote and eukaryote when they are used in a phylogenetic sense. However, they do not constitute a dichotomy; they do not collectively exhaust the class of living systems. There exists a third kingdom which, to date, is represented solely by the methanogenic bacteria, a relatively unknown class of anaerobes that possess a unique metabolism based on the reduction of carbon dioxide to methane (19-21). These bacteria appear to be no more related to typical bacteria than they are to eukaryotic cytoplasms. Although the two divisions of this kingdom appear as remote from one another as blue-green algae are from other eubacteria, they nevertheless correspond to the same biochemical phenotype. The apparent antiquity of the methanogenic phenotype plus the fact that it seems well suited to the type of environment presumed to exist on earth 3-4 billion years ago lead us tentatively to name this urkingdom the archaebacteria. Whether or not other biochemically distinct phenotypes exist in this kingdom is clearly an important question upon which may turn our concept of the nature and ancestry of the first prokaryotes.” Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 64. Propose “three aboriginal lines of descent” ! Eubacteria ! Archaebacteria ! Urkaryotes Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 65. Conclusion “With the identification and characterization of the urkingdoms we are for the first time beginning to see the overall phylogenetic structure of the living world. It is not structured in a bipartite way along the lines of the organizationally dissimilar prokaryote and eukaryote. Rather, it is (at least) tripartite, comprising (i) the typical bacteria, (ii) the line of descent manifested in eukaryotic cytoplasms, and (iii) a little explored grouping, represented so far only by methanogenic bacteria.” Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 66. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 67. Tree from Woese and Fox data http://bioweb.pasteur.fr/seqanal/interfaces/neighbor.html Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 68. Woese 1987 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 69. Woese 1987 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 70. 26.23 Some Would Call It Hell; These Archaea Call It Home Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 70
- 71. Table 26.1 The Three Domains of Life on Earth Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 71
- 72. Table 26.1 The Three Domains of Life on Earth Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 72
- 73. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 74. 26.22 Membrane Architecture in Archaea Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014 74
- 75. Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
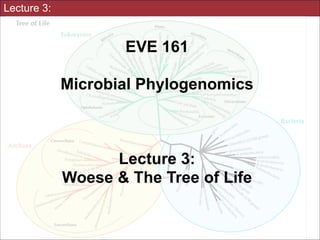
- 76. Tree of Life adapted from Baldauf, et al., in Assembling the Tree of Life, 2004 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 77. Most of the phylogenetic diversity of life is microbial adapted from Baldauf, et al., in Assembling the Tree of Life, 2004 Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 78. Simplified, Rooted Tree of Life Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
- 79. Alternative rooted tree of life Archaea Archaea Slides for UC Davis EVE161 Course Taught by Jonathan Eisen Winter 2014
